Gene
KWMTBOMO08421
Pre Gene Modal
BGIBMGA009301
Annotation
soluble_guanylyl_cyclae_beta-1_subunit_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.514
Sequence
CDS
ATGTCGTTACCGCACACTAACGTTTTGTTTGTGGAAAAGTATGGGTTCGTTAATTACGCTCTTGAGCTGCTCGTGATAAAAACATTTGACGAAGAAACATGGGAGACTATAAAGAAGAAAGCCGACGTAGCGATGGAGGGCAGCTTCCTTGTGCGGCAGATTTATGAAGATGAAATCACGTATAATTTGATCACCGCCGCTGTGGAGGTTCTGCAAATCCCAGCCGACGCGATTTTAGAGTTGTTCGGGAAGACATTCTTTGAGTTCTGCCAAGATTCGGGATACGATAAGATACTTCAAGTACTCGGTGCGACGCCGCGGGACTTCTTACAAAACCTGGACGGGCTACATGATCACTTAGGCACGCTGTACCCTGGCATGCGATCGCCTTCGTTCCGATGCACCGAGAGACCTGAAGACGGGGCTTTGATTCTACATTACTATTCAGATAGGCCAGGGCTAGAACACATCGTCATCGGAATAGTAAAAACCGTAGCAAGCAAGCTTCATAACACGGAAGTCAAAGTGGAAATCTTGAAGACTAAACAGGAATGTGACCACGTCCAGTTTTTGATCACCGAAACTTCTACGACCGGTCGAGTTTCTGCTCCGGAAATTGCTGAGATTGAAACTTTGTCTCTTGAGCCAAAAGTCAGTCCTGCGACATTCTGCCGTGTTTTCCCATTCCATTTGATGTTTGATCGTGAACTGAACATCATTCAAGCTGGAAGGACCGTTTCTCGTCTCCTACCAAGGATCACCAGACCTGGATGCAAGATTACCGACGTTTTGGATACGGTGAGGCCTCATTTGGAAATGACATTCGCTAATGTTCTGGCCCATATCAATACTGTGTACGTGCTGAAGACGAAGCCAGAGGAGATGAGAGCCACCGATCCTCAGGAAGAGATTGCGTCTCTCCGCTTGAAGGGTCAAATGCTCTACATTCCTGAAACGGATGTGGTAGTATTTCAATGCTACCCGAGCGTTACCAACTTGGATGATCTCACGCGTCGTGGACTCTGCATTTCGGACATTCCACTTCATGACGCGACCCGAGATCTCGTACTCATGTCGGAACAGTTCGAAGCTGACTACAAACTAACGCAGAACTTGGAAGTGCTGACCGACAAACTCCAGCAGACCTTCAGGGAACTGGACTCGGAGAAGCAGAAAACGGATAGACTTCTGTACTCAGTTTTGCCGATAAGCGTAGCGACAGAGCTGCGTCATCAACGTCCAGTCCCGGCACGCCGCTATGATCCGGTCACTCTCCTCTTCAGCGGAGTCGTGGGCTTTGCCAACTATTGTGCTAGGAACTCTGACCATAAGGGAGCTATGAAGATTGTGAGGATGCTGAATGACCTGTACACAGCCTTCGATGTTCTCACGGATCCGAAGAGAAATCCCAACGTTTACAAGGTGGAGACAGTAGGGGATAAGTACATGGCGGTGTCTGGTCTGCCGGAGTACGAAGTGGCCCACGCGAAACACATTTCTCTTCTGGCTCTGGACATGATGGATCTGTCCCAAACCGTGACCGTTGATGGAGAGCCTGTGGGTATCACTATAGGCATTCACTCCGGGGAGGTCGTGACAGGTGTGATCGGTCATCGCATGCCTCGTTATTGCCTCTTCGGCAATACGGTAAACCTGACCAGCCGCTGCGAAACTACTGGCGTTCCCGGGACCATTAATGTCAGCGAGGATACATACAACTATCTCATGCGCGATGACAACCACGATGACCAGTTCGAGCTGACGTACCGCGGTCACGTGACCATGAAGGGGAAAGCCGAACCGATGCAAACCTGGTTCCTGACACGGAAGACGATTAATTAA
Protein
MSLPHTNVLFVEKYGFVNYALELLVIKTFDEETWETIKKKADVAMEGSFLVRQIYEDEITYNLITAAVEVLQIPADAILELFGKTFFEFCQDSGYDKILQVLGATPRDFLQNLDGLHDHLGTLYPGMRSPSFRCTERPEDGALILHYYSDRPGLEHIVIGIVKTVASKLHNTEVKVEILKTKQECDHVQFLITETSTTGRVSAPEIAEIETLSLEPKVSPATFCRVFPFHLMFDRELNIIQAGRTVSRLLPRITRPGCKITDVLDTVRPHLEMTFANVLAHINTVYVLKTKPEEMRATDPQEEIASLRLKGQMLYIPETDVVVFQCYPSVTNLDDLTRRGLCISDIPLHDATRDLVLMSEQFEADYKLTQNLEVLTDKLQQTFRELDSEKQKTDRLLYSVLPISVATELRHQRPVPARRYDPVTLLFSGVVGFANYCARNSDHKGAMKIVRMLNDLYTAFDVLTDPKRNPNVYKVETVGDKYMAVSGLPEYEVAHAKHISLLALDMMDLSQTVTVDGEPVGITIGIHSGEVVTGVIGHRMPRYCLFGNTVNLTSRCETTGVPGTINVSEDTYNYLMRDDNHDDQFELTYRGHVTMKGKAEPMQTWFLTRKTIN
Summary
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Uniprot
F8WS78
O77106
A0A3S2PF53
A0A2A4KA63
A0A2H1VV62
A0A0N1I4Y5
+ More
A0A0N0PAS7 A0A2J7R4K5 A0A067QGL7 D1ZZC3 A0A1W4XCJ9 F5HSC0 A0A0M9A423 A0A151JZB9 A0A195E5Y1 A0A1Y1LVV3 A0A0C9RKY3 Q5UAF0 Q6L5L6 J9JNY6 A0A2H8TNN6 A0A2A4KB49 K7J3A7 A0A195CGV5 R4FQM1 A0A0L7R5W5 A0A139WIV2 E0W2A4 E2BB36 A0A0A9X0F7 A0A2P8Z3U1 E2AP12 A0A026WGQ5 A0A226DEV9 B7PSP7 A0A154PR70 T1KFI6 A0A1D2NMJ1 A0A158N9T7 A2SW27 T1ITQ7 M9WNL3 Q59HN5 A0A3L8DR43 A0A1Y1LXH4 C3XW68 E9FT54 A0A0P5PVF0 A0A0P5UHJ2 A2SW28 F4WJW3 K1S3W9 B2MVM1 A0A2C9JHL9 A0A0P6HD79 A0A0B6ZPJ3 A0A0N8DK31 A0A210QJM4 A0A0T6BGM9 A0A401T2I7 W5MNT1 A0A3Q0FU18 V4CJ11 A0A091UI47 A0A091RI65 A0A1U7SIT8 A0A0P7YXV8 A0A093F6U6 A0A218UD84 A0A3M0KLC2 A0A1W4YT06 A0A1V4JTH5 A0A093I423 A0A315W0B2 A0A3Q2E1I5 A0A091HX37 A0A1V4JT08 A0A3Q2CUP1 A0A1D5PAM2 A0A1D5PG10 A0A452H986 W5UBV5 A0A452H944 A0A3B3THL6 A0A452H982 A0A3P9NCK2 A0A3B3XMH1 A0A3Q1IY71 A0A087XAN5 A0A1A8RI55 A0A1A8IGG1 A0A1A8QNN8 A0A1A8CQB8 A0A1A8F2M3 A0A1A7ZBA7 M3ZU00 A0A3P8XFG8 A0A3B4WVP7 A0A1W4YIP9 A0A3B4TD52
A0A0N0PAS7 A0A2J7R4K5 A0A067QGL7 D1ZZC3 A0A1W4XCJ9 F5HSC0 A0A0M9A423 A0A151JZB9 A0A195E5Y1 A0A1Y1LVV3 A0A0C9RKY3 Q5UAF0 Q6L5L6 J9JNY6 A0A2H8TNN6 A0A2A4KB49 K7J3A7 A0A195CGV5 R4FQM1 A0A0L7R5W5 A0A139WIV2 E0W2A4 E2BB36 A0A0A9X0F7 A0A2P8Z3U1 E2AP12 A0A026WGQ5 A0A226DEV9 B7PSP7 A0A154PR70 T1KFI6 A0A1D2NMJ1 A0A158N9T7 A2SW27 T1ITQ7 M9WNL3 Q59HN5 A0A3L8DR43 A0A1Y1LXH4 C3XW68 E9FT54 A0A0P5PVF0 A0A0P5UHJ2 A2SW28 F4WJW3 K1S3W9 B2MVM1 A0A2C9JHL9 A0A0P6HD79 A0A0B6ZPJ3 A0A0N8DK31 A0A210QJM4 A0A0T6BGM9 A0A401T2I7 W5MNT1 A0A3Q0FU18 V4CJ11 A0A091UI47 A0A091RI65 A0A1U7SIT8 A0A0P7YXV8 A0A093F6U6 A0A218UD84 A0A3M0KLC2 A0A1W4YT06 A0A1V4JTH5 A0A093I423 A0A315W0B2 A0A3Q2E1I5 A0A091HX37 A0A1V4JT08 A0A3Q2CUP1 A0A1D5PAM2 A0A1D5PG10 A0A452H986 W5UBV5 A0A452H944 A0A3B3THL6 A0A452H982 A0A3P9NCK2 A0A3B3XMH1 A0A3Q1IY71 A0A087XAN5 A0A1A8RI55 A0A1A8IGG1 A0A1A8QNN8 A0A1A8CQB8 A0A1A8F2M3 A0A1A7ZBA7 M3ZU00 A0A3P8XFG8 A0A3B4WVP7 A0A1W4YIP9 A0A3B4TD52
Pubmed
EMBL
AB645767
BAK52271.1
AF062751
AAC61264.1
RSAL01000059
RVE49670.1
+ More
NWSH01000007 PCG80969.1 ODYU01004643 SOQ44735.1 KQ459256 KPJ02076.1 KQ461161 KPJ08006.1 NEVH01007407 PNF35756.1 KK853463 KDR07449.1 KQ971338 EFA01870.2 AB630171 BAK23251.1 KQ435740 KOX76795.1 KQ981404 KYN41942.1 KQ979592 KYN20605.1 GEZM01049952 JAV75736.1 GBYB01004006 GBYB01008850 JAG73773.1 JAG78617.1 AY769960 AAV34676.1 AB181489 BAD22772.1 ABLF02026149 ABLF02026150 GFXV01003834 MBW15639.1 PCG80970.1 KQ977791 KYM99932.1 GAHY01000324 JAA77186.1 KQ414648 KOC66209.1 KYB27920.1 DS235875 EEB19760.1 GL446901 EFN87127.1 GBHO01030468 GDHC01015713 JAG13136.1 JAQ02916.1 PYGN01000209 PSN51153.1 GL441439 EFN64844.1 KK107214 EZA55247.1 LNIX01000020 OXA44092.1 ABJB010292149 ABJB010652590 ABJB010818711 ABJB011091018 DS779769 EEC09619.1 KQ435066 KZC14381.1 CAEY01000037 LJIJ01000004 ODN06498.1 ADTU01009520 ADTU01009521 ADTU01009522 ADTU01009523 ADTU01009524 ADTU01009525 ADTU01009526 ADTU01009527 DQ355433 ABC94528.1 JH431494 KC495565 AGJ94942.1 AB207898 BAD91320.1 QOIP01000005 RLU22877.1 GEZM01049951 JAV75737.1 GG666471 EEN67602.1 GL732524 EFX89313.1 GDIQ01133265 JAL18461.1 GDIP01112837 JAL90877.1 DQ355434 ABC94529.1 GL888187 EGI65485.1 JH816850 EKC42091.1 EU624340 ACC95432.1 GDIQ01020926 JAN73811.1 HACG01023593 CEK70458.1 GDIP01025999 JAM77716.1 NEDP02003345 OWF48957.1 LJIG01000460 KRT86480.1 BEZZ01000893 GCC36838.1 AHAT01015360 AHAT01015361 AHAT01015362 KB200329 ESP02185.1 KK452892 KFQ74643.1 KK817856 KFQ38947.1 JARO02002304 KPP73018.1 KK387611 KFV52205.1 MUZQ01000442 OWK51362.1 QRBI01000105 RMC13865.1 LSYS01006437 OPJ75345.1 KL214744 KFV61521.1 NHOQ01000621 PWA29374.1 KL217999 KFP00854.1 OPJ75346.1 AADN05000018 JT411577 AHH39501.1 AYCK01004554 HAEH01004135 HAEI01008714 SBS05775.1 HAED01009822 HAEE01006024 SBQ96034.1 HAEF01007830 HAEG01013494 SBR95137.1 HADZ01018026 HAEA01004616 SBP81967.1 HAEB01007063 HAEC01011775 SBQ53590.1 HADY01001111 HAEJ01008413 SBP39596.1
NWSH01000007 PCG80969.1 ODYU01004643 SOQ44735.1 KQ459256 KPJ02076.1 KQ461161 KPJ08006.1 NEVH01007407 PNF35756.1 KK853463 KDR07449.1 KQ971338 EFA01870.2 AB630171 BAK23251.1 KQ435740 KOX76795.1 KQ981404 KYN41942.1 KQ979592 KYN20605.1 GEZM01049952 JAV75736.1 GBYB01004006 GBYB01008850 JAG73773.1 JAG78617.1 AY769960 AAV34676.1 AB181489 BAD22772.1 ABLF02026149 ABLF02026150 GFXV01003834 MBW15639.1 PCG80970.1 KQ977791 KYM99932.1 GAHY01000324 JAA77186.1 KQ414648 KOC66209.1 KYB27920.1 DS235875 EEB19760.1 GL446901 EFN87127.1 GBHO01030468 GDHC01015713 JAG13136.1 JAQ02916.1 PYGN01000209 PSN51153.1 GL441439 EFN64844.1 KK107214 EZA55247.1 LNIX01000020 OXA44092.1 ABJB010292149 ABJB010652590 ABJB010818711 ABJB011091018 DS779769 EEC09619.1 KQ435066 KZC14381.1 CAEY01000037 LJIJ01000004 ODN06498.1 ADTU01009520 ADTU01009521 ADTU01009522 ADTU01009523 ADTU01009524 ADTU01009525 ADTU01009526 ADTU01009527 DQ355433 ABC94528.1 JH431494 KC495565 AGJ94942.1 AB207898 BAD91320.1 QOIP01000005 RLU22877.1 GEZM01049951 JAV75737.1 GG666471 EEN67602.1 GL732524 EFX89313.1 GDIQ01133265 JAL18461.1 GDIP01112837 JAL90877.1 DQ355434 ABC94529.1 GL888187 EGI65485.1 JH816850 EKC42091.1 EU624340 ACC95432.1 GDIQ01020926 JAN73811.1 HACG01023593 CEK70458.1 GDIP01025999 JAM77716.1 NEDP02003345 OWF48957.1 LJIG01000460 KRT86480.1 BEZZ01000893 GCC36838.1 AHAT01015360 AHAT01015361 AHAT01015362 KB200329 ESP02185.1 KK452892 KFQ74643.1 KK817856 KFQ38947.1 JARO02002304 KPP73018.1 KK387611 KFV52205.1 MUZQ01000442 OWK51362.1 QRBI01000105 RMC13865.1 LSYS01006437 OPJ75345.1 KL214744 KFV61521.1 NHOQ01000621 PWA29374.1 KL217999 KFP00854.1 OPJ75346.1 AADN05000018 JT411577 AHH39501.1 AYCK01004554 HAEH01004135 HAEI01008714 SBS05775.1 HAED01009822 HAEE01006024 SBQ96034.1 HAEF01007830 HAEG01013494 SBR95137.1 HADZ01018026 HAEA01004616 SBP81967.1 HAEB01007063 HAEC01011775 SBQ53590.1 HADY01001111 HAEJ01008413 SBP39596.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000235965
UP000027135
+ More
UP000007266 UP000192223 UP000053105 UP000078541 UP000078492 UP000005203 UP000007819 UP000002358 UP000078542 UP000053825 UP000009046 UP000008237 UP000245037 UP000000311 UP000053097 UP000198287 UP000001555 UP000076502 UP000015104 UP000094527 UP000005205 UP000279307 UP000001554 UP000000305 UP000007755 UP000005408 UP000076420 UP000242188 UP000287033 UP000018468 UP000189705 UP000030746 UP000034805 UP000197619 UP000269221 UP000192224 UP000190648 UP000053875 UP000265020 UP000054308 UP000000539 UP000291020 UP000221080 UP000261540 UP000242638 UP000261480 UP000265040 UP000028760 UP000002852 UP000265140 UP000261360 UP000261420
UP000007266 UP000192223 UP000053105 UP000078541 UP000078492 UP000005203 UP000007819 UP000002358 UP000078542 UP000053825 UP000009046 UP000008237 UP000245037 UP000000311 UP000053097 UP000198287 UP000001555 UP000076502 UP000015104 UP000094527 UP000005205 UP000279307 UP000001554 UP000000305 UP000007755 UP000005408 UP000076420 UP000242188 UP000287033 UP000018468 UP000189705 UP000030746 UP000034805 UP000197619 UP000269221 UP000192224 UP000190648 UP000053875 UP000265020 UP000054308 UP000000539 UP000291020 UP000221080 UP000261540 UP000242638 UP000261480 UP000265040 UP000028760 UP000002852 UP000265140 UP000261360 UP000261420
PRIDE
Interpro
Gene 3D
ProteinModelPortal
F8WS78
O77106
A0A3S2PF53
A0A2A4KA63
A0A2H1VV62
A0A0N1I4Y5
+ More
A0A0N0PAS7 A0A2J7R4K5 A0A067QGL7 D1ZZC3 A0A1W4XCJ9 F5HSC0 A0A0M9A423 A0A151JZB9 A0A195E5Y1 A0A1Y1LVV3 A0A0C9RKY3 Q5UAF0 Q6L5L6 J9JNY6 A0A2H8TNN6 A0A2A4KB49 K7J3A7 A0A195CGV5 R4FQM1 A0A0L7R5W5 A0A139WIV2 E0W2A4 E2BB36 A0A0A9X0F7 A0A2P8Z3U1 E2AP12 A0A026WGQ5 A0A226DEV9 B7PSP7 A0A154PR70 T1KFI6 A0A1D2NMJ1 A0A158N9T7 A2SW27 T1ITQ7 M9WNL3 Q59HN5 A0A3L8DR43 A0A1Y1LXH4 C3XW68 E9FT54 A0A0P5PVF0 A0A0P5UHJ2 A2SW28 F4WJW3 K1S3W9 B2MVM1 A0A2C9JHL9 A0A0P6HD79 A0A0B6ZPJ3 A0A0N8DK31 A0A210QJM4 A0A0T6BGM9 A0A401T2I7 W5MNT1 A0A3Q0FU18 V4CJ11 A0A091UI47 A0A091RI65 A0A1U7SIT8 A0A0P7YXV8 A0A093F6U6 A0A218UD84 A0A3M0KLC2 A0A1W4YT06 A0A1V4JTH5 A0A093I423 A0A315W0B2 A0A3Q2E1I5 A0A091HX37 A0A1V4JT08 A0A3Q2CUP1 A0A1D5PAM2 A0A1D5PG10 A0A452H986 W5UBV5 A0A452H944 A0A3B3THL6 A0A452H982 A0A3P9NCK2 A0A3B3XMH1 A0A3Q1IY71 A0A087XAN5 A0A1A8RI55 A0A1A8IGG1 A0A1A8QNN8 A0A1A8CQB8 A0A1A8F2M3 A0A1A7ZBA7 M3ZU00 A0A3P8XFG8 A0A3B4WVP7 A0A1W4YIP9 A0A3B4TD52
A0A0N0PAS7 A0A2J7R4K5 A0A067QGL7 D1ZZC3 A0A1W4XCJ9 F5HSC0 A0A0M9A423 A0A151JZB9 A0A195E5Y1 A0A1Y1LVV3 A0A0C9RKY3 Q5UAF0 Q6L5L6 J9JNY6 A0A2H8TNN6 A0A2A4KB49 K7J3A7 A0A195CGV5 R4FQM1 A0A0L7R5W5 A0A139WIV2 E0W2A4 E2BB36 A0A0A9X0F7 A0A2P8Z3U1 E2AP12 A0A026WGQ5 A0A226DEV9 B7PSP7 A0A154PR70 T1KFI6 A0A1D2NMJ1 A0A158N9T7 A2SW27 T1ITQ7 M9WNL3 Q59HN5 A0A3L8DR43 A0A1Y1LXH4 C3XW68 E9FT54 A0A0P5PVF0 A0A0P5UHJ2 A2SW28 F4WJW3 K1S3W9 B2MVM1 A0A2C9JHL9 A0A0P6HD79 A0A0B6ZPJ3 A0A0N8DK31 A0A210QJM4 A0A0T6BGM9 A0A401T2I7 W5MNT1 A0A3Q0FU18 V4CJ11 A0A091UI47 A0A091RI65 A0A1U7SIT8 A0A0P7YXV8 A0A093F6U6 A0A218UD84 A0A3M0KLC2 A0A1W4YT06 A0A1V4JTH5 A0A093I423 A0A315W0B2 A0A3Q2E1I5 A0A091HX37 A0A1V4JT08 A0A3Q2CUP1 A0A1D5PAM2 A0A1D5PG10 A0A452H986 W5UBV5 A0A452H944 A0A3B3THL6 A0A452H982 A0A3P9NCK2 A0A3B3XMH1 A0A3Q1IY71 A0A087XAN5 A0A1A8RI55 A0A1A8IGG1 A0A1A8QNN8 A0A1A8CQB8 A0A1A8F2M3 A0A1A7ZBA7 M3ZU00 A0A3P8XFG8 A0A3B4WVP7 A0A1W4YIP9 A0A3B4TD52
PDB
4NI2
E-value=3.68108e-78,
Score=744
Ontologies
PATHWAY
GO
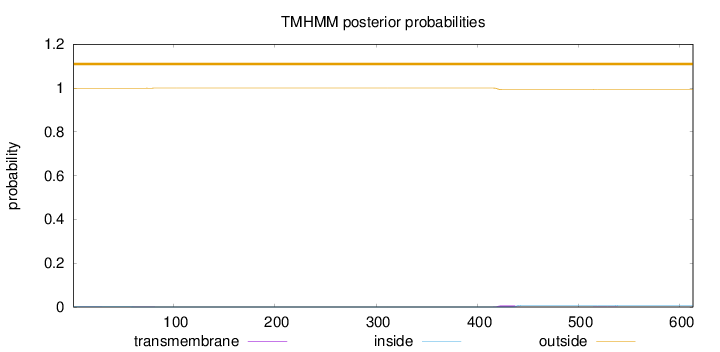
Topology
Length:
613
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16306
Exp number, first 60 AAs:
0.01075
Total prob of N-in:
0.00108
outside
1 - 613
Population Genetic Test Statistics
Pi
340.079166
Theta
177.231482
Tajima's D
1.304372
CLR
0.160872
CSRT
0.741862906854657
Interpretation
Uncertain
