Gene
KWMTBOMO08415
Pre Gene Modal
BGIBMGA009300
Annotation
PREDICTED:_soluble_guanylyl_cyclae_alpha-1_subunit_isoform_X1_[Bombyx_mori]
Full name
Head-specific guanylate cyclase
Alternative Name
Gycalpha99B
Location in the cell
Cytoplasmic Reliability : 3.163
Sequence
CDS
ATGGTGTTCCCTATGTTCTGTTTTCTCCTACAGGGCTTAGAAATAAAGGGCCAAATGGTGTTTTGTTCCGAATCGGATTCCCTTCTCTTCGTGGGCTCTCCGTTCCTTGATGGACTTGAAGGCTTAACTGGGAGAGGCCTTTTCATATCAGATATTCCACTCCACGATGCAACAAGGGATGTCATTCTAGTGGGAGAACAGGCTCGGGCACAGGACGGACTTCGAAGGCGTATGGACAAACTTAAGAACTCAATCGAAGAAGCAAGTAAAGCGGTAGACAAAGAGCGCGAGAAGAATGTCAGTCTTCTGCATTTAATATTCCCACCTCACATCGCTAAAAGGCTGTGGCTAGGTGAGAAGATAGAAGCGAAAAGCCACGATGACGTCACAATGCTGTTCAGTGACATCGTCGGCTTCACATCCATCTGTGCTACTGCAACACCGATGATGGTTATTGCGATGCTGGAGGATCTGTACAGCGTATTTGATATATTCTGTGAGGAGCTTGACGTTTATAAGGTGGAAACAATAGGTGATGCATATTGCGTTGCAAGTGGCTTACACCGCAAAATCGAGACGCACGCACCCCAGATCGCATGGATGGCGTTACGCATGGTCGAAACATGCGCACAGCATTTGACGCATGAAGGAAATCCCATCAAGATGCGCTTAGGACTGCATACTGGTACAGTACTAGCCGGGGTCGTCGGTAAAACTATGCAGAAATACTGCCTTTTCGGTCATAACGTGACCCTTGCAAATAAATTTGAATCCGGCTCAGAACCACTGAAGATCAACGTCAGTCCCACCACTTACGAGTGGCTAATACAATTCCCGGGCTTCGAAATGGAATCGCGAGACCGTTCCTGTCTCCCGAATTCATTCCCCAAGGACATCCCGGGAACATGTTATTTCCTTCACAAGTACGTACACCCGGGAACGGACCCTGCGGCGTCCCAGGTCAAACATATACAGGATGCGTTAAAATATTATGAAATAGGACAATTCGGACAAAGTAATCCAGTTGAAAATGACCCGGAGCCAACATAG
Protein
MVFPMFCFLLQGLEIKGQMVFCSESDSLLFVGSPFLDGLEGLTGRGLFISDIPLHDATRDVILVGEQARAQDGLRRRMDKLKNSIEEASKAVDKEREKNVSLLHLIFPPHIAKRLWLGEKIEAKSHDDVTMLFSDIVGFTSICATATPMMVIAMLEDLYSVFDIFCEELDVYKVETIGDAYCVASGLHRKIETHAPQIAWMALRMVETCAQHLTHEGNPIKMRLGLHTGTVLAGVVGKTMQKYCLFGHNVTLANKFESGSEPLKINVSPTTYEWLIQFPGFEMESRDRSCLPNSFPKDIPGTCYFLHKYVHPGTDPAASQVKHIQDALKYYEIGQFGQSNPVENDPEPT
Summary
Description
May have a role in phototransduction. A second subunit may be required for enzyme activity.
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Subunit
Dimer.
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Keywords
cGMP biosynthesis
Complete proteome
Cytoplasm
GTP-binding
Lyase
Nucleotide-binding
Reference proteome
Sensory transduction
Vision
Feature
chain Head-specific guanylate cyclase
Uniprot
H9JIF0
A0A2A4K9R7
A0A0N0PA12
A0A0N1I9V6
O77105
A0A437BH12
+ More
F8WS79 A0A212EUS7 A0A0L7LB52 D1ZZC5 F5HSB9 A0A067QTW8 A0A1Y1KL30 A0A1Y1KNE3 A0A1B6HW21 A0A0C9RY60 A0A1B6F0N5 A0A232EH27 K7J3A4 A0A026WHV8 A0A1D2NMJ9 A0A1S4E8I9 E2BB35 E2AP14 A0A3L8DQ36 A0A158N9E4 A0A2S2PET4 F4WJW0 A0A2H8TI48 J9K5C7 A0A151JZH4 T1HN04 A0A195CGS2 A0A2S2Q520 A0A088AMV1 Q5W7P2 A0A2A3ES47 E9FT53 A0A195BGN4 A0A154PR62 A0A151XI26 A0A195E615 W8B6M7 A0A1A9WB53 A0A0L0CEM3 Q7YZQ2 A0A0K8VIW2 A0A034WES2 A0A0A1XFX4 A0A1A9VYA9 A0A0P5KT04 A0A1B6CQ32 A0A164LKH5 A0A1A9Y948 A0A1B0AGI7 B4NBJ6 A0A1B0FR75 A0A1I8Q0Y3 A0A1I8N3V3 A0A084VNW7 A0A182JIE4 A0A182QMV2 T1P7Y8 A0A182MG85 A0A182N8F9 A0A182PSU5 W5JKG2 A0A182FIT2 B0WSU5 A0A182KEJ3 Q17L76 A0A182W5M0 A0A182T0S2 A0A0L7R5P2 A0A1S4EYY2 A0A182TFT6 A0A182XBY3 F5HJM4 A0A182UNC6 Q7QF31 A0A182IAK2 A0A182HD00 A0A1Q3FZX7 A0A182RHK6 A0A1B0GK38 A0A182KLP8 A0A0P6H817 A0A182YH65 A0A1B0C539 A0A147BLC7 A0A0M4EI23 B3P5F2 B4R067 A0A3B0JKP3 A0A1W4UPX8 B4PPM5 B3MTG6 H5V8C6 Q07093
F8WS79 A0A212EUS7 A0A0L7LB52 D1ZZC5 F5HSB9 A0A067QTW8 A0A1Y1KL30 A0A1Y1KNE3 A0A1B6HW21 A0A0C9RY60 A0A1B6F0N5 A0A232EH27 K7J3A4 A0A026WHV8 A0A1D2NMJ9 A0A1S4E8I9 E2BB35 E2AP14 A0A3L8DQ36 A0A158N9E4 A0A2S2PET4 F4WJW0 A0A2H8TI48 J9K5C7 A0A151JZH4 T1HN04 A0A195CGS2 A0A2S2Q520 A0A088AMV1 Q5W7P2 A0A2A3ES47 E9FT53 A0A195BGN4 A0A154PR62 A0A151XI26 A0A195E615 W8B6M7 A0A1A9WB53 A0A0L0CEM3 Q7YZQ2 A0A0K8VIW2 A0A034WES2 A0A0A1XFX4 A0A1A9VYA9 A0A0P5KT04 A0A1B6CQ32 A0A164LKH5 A0A1A9Y948 A0A1B0AGI7 B4NBJ6 A0A1B0FR75 A0A1I8Q0Y3 A0A1I8N3V3 A0A084VNW7 A0A182JIE4 A0A182QMV2 T1P7Y8 A0A182MG85 A0A182N8F9 A0A182PSU5 W5JKG2 A0A182FIT2 B0WSU5 A0A182KEJ3 Q17L76 A0A182W5M0 A0A182T0S2 A0A0L7R5P2 A0A1S4EYY2 A0A182TFT6 A0A182XBY3 F5HJM4 A0A182UNC6 Q7QF31 A0A182IAK2 A0A182HD00 A0A1Q3FZX7 A0A182RHK6 A0A1B0GK38 A0A182KLP8 A0A0P6H817 A0A182YH65 A0A1B0C539 A0A147BLC7 A0A0M4EI23 B3P5F2 B4R067 A0A3B0JKP3 A0A1W4UPX8 B4PPM5 B3MTG6 H5V8C6 Q07093
EC Number
4.6.1.2
Pubmed
19121390
26354079
9736646
23934793
22118469
26227816
+ More
18362917 19820115 24845553 28004739 28648823 20075255 24508170 27289101 20798317 30249741 21347285 21719571 21292972 24495485 26108605 25348373 25830018 17994087 25315136 24438588 20920257 23761445 17510324 12364791 14747013 17210077 26483478 20966253 25244985 29652888 17550304 8095978 7797526 10731132 12537572 12537569
18362917 19820115 24845553 28004739 28648823 20075255 24508170 27289101 20798317 30249741 21347285 21719571 21292972 24495485 26108605 25348373 25830018 17994087 25315136 24438588 20920257 23761445 17510324 12364791 14747013 17210077 26483478 20966253 25244985 29652888 17550304 8095978 7797526 10731132 12537572 12537569
EMBL
BABH01022496
BABH01022497
BABH01022498
NWSH01000007
PCG80971.1
KQ459256
+ More
KPJ02078.1 KQ460472 KPJ14572.1 AF062750 AAC61263.1 RSAL01000059 RVE49671.1 AB645768 BAK52272.1 AGBW02012298 OWR45245.1 JTDY01001858 KOB72703.1 KQ971338 EFA02348.1 AB630170 BAK23250.1 KK853463 KDR07447.1 GEZM01084965 JAV60365.1 GEZM01084963 JAV60367.1 GECU01028870 JAS78836.1 GBYB01014205 JAG83972.1 GECZ01025950 JAS43819.1 NNAY01004604 OXU17667.1 KK107214 EZA55251.1 LJIJ01000004 ODN06500.1 GL446901 EFN87126.1 GL441439 EFN64846.1 QOIP01000005 RLU22481.1 ADTU01009527 ADTU01009528 GGMR01015296 MBY27915.1 GL888187 EGI65482.1 GFXV01001914 MBW13719.1 ABLF02027805 ABLF02027806 ABLF02027807 ABLF02027808 ABLF02027809 ABLF02029742 ABLF02029743 ABLF02029745 KQ981404 KYN41941.1 ACPB03002869 KQ977791 KYM99935.1 GGMS01003625 MBY72828.1 AB193550 BAD66824.1 KZ288190 PBC34527.1 GL732524 EFX89707.1 KQ976476 KYM83831.1 KQ435066 KZC14382.1 KQ982093 KYQ60049.1 KQ979592 KYN20608.1 GAMC01013862 JAB92693.1 JRES01000501 KNC30705.1 AY324653 AAP85539.1 GDHF01013483 JAI38831.1 GAKP01004871 JAC54081.1 GBXI01004446 JAD09846.1 GDIQ01185249 JAK66476.1 GEDC01021686 JAS15612.1 LRGB01003123 KZS04198.1 CH964232 EDW81160.1 CCAG010005441 ATLV01014933 KE524996 KFB39661.1 AXCN02002153 KA644667 AFP59296.1 AXCM01005054 ADMH02001274 ETN63264.1 DS232076 EDS34055.1 CH477218 EAT47419.1 KQ414648 KOC66207.1 AAAB01008846 EGK96485.1 EAA06502.5 APCN01001259 JXUM01034566 KQ561028 KXJ79976.1 GFDL01001965 JAV33080.1 AJWK01022543 GDIQ01023305 JAN71432.1 JXJN01025806 JXJN01025807 JXJN01025808 JXJN01025809 JXJN01025810 GEGO01003811 JAR91593.1 CP012526 ALC46196.1 CH954182 EDV53202.1 CM000364 EDX14873.1 OUUW01000007 SPP82897.1 CM000160 EDW98275.1 CH902623 EDV30556.1 BT133227 AFA36410.1 S57126 U27117 AE014297 AY060654
KPJ02078.1 KQ460472 KPJ14572.1 AF062750 AAC61263.1 RSAL01000059 RVE49671.1 AB645768 BAK52272.1 AGBW02012298 OWR45245.1 JTDY01001858 KOB72703.1 KQ971338 EFA02348.1 AB630170 BAK23250.1 KK853463 KDR07447.1 GEZM01084965 JAV60365.1 GEZM01084963 JAV60367.1 GECU01028870 JAS78836.1 GBYB01014205 JAG83972.1 GECZ01025950 JAS43819.1 NNAY01004604 OXU17667.1 KK107214 EZA55251.1 LJIJ01000004 ODN06500.1 GL446901 EFN87126.1 GL441439 EFN64846.1 QOIP01000005 RLU22481.1 ADTU01009527 ADTU01009528 GGMR01015296 MBY27915.1 GL888187 EGI65482.1 GFXV01001914 MBW13719.1 ABLF02027805 ABLF02027806 ABLF02027807 ABLF02027808 ABLF02027809 ABLF02029742 ABLF02029743 ABLF02029745 KQ981404 KYN41941.1 ACPB03002869 KQ977791 KYM99935.1 GGMS01003625 MBY72828.1 AB193550 BAD66824.1 KZ288190 PBC34527.1 GL732524 EFX89707.1 KQ976476 KYM83831.1 KQ435066 KZC14382.1 KQ982093 KYQ60049.1 KQ979592 KYN20608.1 GAMC01013862 JAB92693.1 JRES01000501 KNC30705.1 AY324653 AAP85539.1 GDHF01013483 JAI38831.1 GAKP01004871 JAC54081.1 GBXI01004446 JAD09846.1 GDIQ01185249 JAK66476.1 GEDC01021686 JAS15612.1 LRGB01003123 KZS04198.1 CH964232 EDW81160.1 CCAG010005441 ATLV01014933 KE524996 KFB39661.1 AXCN02002153 KA644667 AFP59296.1 AXCM01005054 ADMH02001274 ETN63264.1 DS232076 EDS34055.1 CH477218 EAT47419.1 KQ414648 KOC66207.1 AAAB01008846 EGK96485.1 EAA06502.5 APCN01001259 JXUM01034566 KQ561028 KXJ79976.1 GFDL01001965 JAV33080.1 AJWK01022543 GDIQ01023305 JAN71432.1 JXJN01025806 JXJN01025807 JXJN01025808 JXJN01025809 JXJN01025810 GEGO01003811 JAR91593.1 CP012526 ALC46196.1 CH954182 EDV53202.1 CM000364 EDX14873.1 OUUW01000007 SPP82897.1 CM000160 EDW98275.1 CH902623 EDV30556.1 BT133227 AFA36410.1 S57126 U27117 AE014297 AY060654
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000037510 UP000007266 UP000027135 UP000215335 UP000002358 UP000053097 UP000094527 UP000079169 UP000008237 UP000000311 UP000279307 UP000005205 UP000007755 UP000007819 UP000078541 UP000015103 UP000078542 UP000005203 UP000242457 UP000000305 UP000078540 UP000076502 UP000075809 UP000078492 UP000091820 UP000037069 UP000078200 UP000076858 UP000092443 UP000092445 UP000007798 UP000092444 UP000095300 UP000095301 UP000030765 UP000075880 UP000075886 UP000075883 UP000075884 UP000075885 UP000000673 UP000069272 UP000002320 UP000075881 UP000008820 UP000075920 UP000075901 UP000053825 UP000075902 UP000076407 UP000007062 UP000075903 UP000075840 UP000069940 UP000249989 UP000075900 UP000092461 UP000075882 UP000076408 UP000092460 UP000092553 UP000008711 UP000000304 UP000268350 UP000192221 UP000002282 UP000007801 UP000000803
UP000037510 UP000007266 UP000027135 UP000215335 UP000002358 UP000053097 UP000094527 UP000079169 UP000008237 UP000000311 UP000279307 UP000005205 UP000007755 UP000007819 UP000078541 UP000015103 UP000078542 UP000005203 UP000242457 UP000000305 UP000078540 UP000076502 UP000075809 UP000078492 UP000091820 UP000037069 UP000078200 UP000076858 UP000092443 UP000092445 UP000007798 UP000092444 UP000095300 UP000095301 UP000030765 UP000075880 UP000075886 UP000075883 UP000075884 UP000075885 UP000000673 UP000069272 UP000002320 UP000075881 UP000008820 UP000075920 UP000075901 UP000053825 UP000075902 UP000076407 UP000007062 UP000075903 UP000075840 UP000069940 UP000249989 UP000075900 UP000092461 UP000075882 UP000076408 UP000092460 UP000092553 UP000008711 UP000000304 UP000268350 UP000192221 UP000002282 UP000007801 UP000000803
Interpro
IPR001054
A/G_cyclase
+ More
IPR011645 HNOB_dom_associated
IPR029787 Nucleotide_cyclase
IPR018297 A/G_cyclase_CS
IPR038158 H-NOX_domain_sf
IPR024096 NO_sig/Golgi_transp_ligand-bd
IPR011644 Heme_NO-bd
IPR004843 Calcineurin-like_PHP_ApaH
IPR008139 SaposinB_dom
IPR029052 Metallo-depent_PP-like
IPR011001 Saposin-like
IPR041805 ASMase/PPN1_MPP
IPR011645 HNOB_dom_associated
IPR029787 Nucleotide_cyclase
IPR018297 A/G_cyclase_CS
IPR038158 H-NOX_domain_sf
IPR024096 NO_sig/Golgi_transp_ligand-bd
IPR011644 Heme_NO-bd
IPR004843 Calcineurin-like_PHP_ApaH
IPR008139 SaposinB_dom
IPR029052 Metallo-depent_PP-like
IPR011001 Saposin-like
IPR041805 ASMase/PPN1_MPP
Gene 3D
ProteinModelPortal
H9JIF0
A0A2A4K9R7
A0A0N0PA12
A0A0N1I9V6
O77105
A0A437BH12
+ More
F8WS79 A0A212EUS7 A0A0L7LB52 D1ZZC5 F5HSB9 A0A067QTW8 A0A1Y1KL30 A0A1Y1KNE3 A0A1B6HW21 A0A0C9RY60 A0A1B6F0N5 A0A232EH27 K7J3A4 A0A026WHV8 A0A1D2NMJ9 A0A1S4E8I9 E2BB35 E2AP14 A0A3L8DQ36 A0A158N9E4 A0A2S2PET4 F4WJW0 A0A2H8TI48 J9K5C7 A0A151JZH4 T1HN04 A0A195CGS2 A0A2S2Q520 A0A088AMV1 Q5W7P2 A0A2A3ES47 E9FT53 A0A195BGN4 A0A154PR62 A0A151XI26 A0A195E615 W8B6M7 A0A1A9WB53 A0A0L0CEM3 Q7YZQ2 A0A0K8VIW2 A0A034WES2 A0A0A1XFX4 A0A1A9VYA9 A0A0P5KT04 A0A1B6CQ32 A0A164LKH5 A0A1A9Y948 A0A1B0AGI7 B4NBJ6 A0A1B0FR75 A0A1I8Q0Y3 A0A1I8N3V3 A0A084VNW7 A0A182JIE4 A0A182QMV2 T1P7Y8 A0A182MG85 A0A182N8F9 A0A182PSU5 W5JKG2 A0A182FIT2 B0WSU5 A0A182KEJ3 Q17L76 A0A182W5M0 A0A182T0S2 A0A0L7R5P2 A0A1S4EYY2 A0A182TFT6 A0A182XBY3 F5HJM4 A0A182UNC6 Q7QF31 A0A182IAK2 A0A182HD00 A0A1Q3FZX7 A0A182RHK6 A0A1B0GK38 A0A182KLP8 A0A0P6H817 A0A182YH65 A0A1B0C539 A0A147BLC7 A0A0M4EI23 B3P5F2 B4R067 A0A3B0JKP3 A0A1W4UPX8 B4PPM5 B3MTG6 H5V8C6 Q07093
F8WS79 A0A212EUS7 A0A0L7LB52 D1ZZC5 F5HSB9 A0A067QTW8 A0A1Y1KL30 A0A1Y1KNE3 A0A1B6HW21 A0A0C9RY60 A0A1B6F0N5 A0A232EH27 K7J3A4 A0A026WHV8 A0A1D2NMJ9 A0A1S4E8I9 E2BB35 E2AP14 A0A3L8DQ36 A0A158N9E4 A0A2S2PET4 F4WJW0 A0A2H8TI48 J9K5C7 A0A151JZH4 T1HN04 A0A195CGS2 A0A2S2Q520 A0A088AMV1 Q5W7P2 A0A2A3ES47 E9FT53 A0A195BGN4 A0A154PR62 A0A151XI26 A0A195E615 W8B6M7 A0A1A9WB53 A0A0L0CEM3 Q7YZQ2 A0A0K8VIW2 A0A034WES2 A0A0A1XFX4 A0A1A9VYA9 A0A0P5KT04 A0A1B6CQ32 A0A164LKH5 A0A1A9Y948 A0A1B0AGI7 B4NBJ6 A0A1B0FR75 A0A1I8Q0Y3 A0A1I8N3V3 A0A084VNW7 A0A182JIE4 A0A182QMV2 T1P7Y8 A0A182MG85 A0A182N8F9 A0A182PSU5 W5JKG2 A0A182FIT2 B0WSU5 A0A182KEJ3 Q17L76 A0A182W5M0 A0A182T0S2 A0A0L7R5P2 A0A1S4EYY2 A0A182TFT6 A0A182XBY3 F5HJM4 A0A182UNC6 Q7QF31 A0A182IAK2 A0A182HD00 A0A1Q3FZX7 A0A182RHK6 A0A1B0GK38 A0A182KLP8 A0A0P6H817 A0A182YH65 A0A1B0C539 A0A147BLC7 A0A0M4EI23 B3P5F2 B4R067 A0A3B0JKP3 A0A1W4UPX8 B4PPM5 B3MTG6 H5V8C6 Q07093
PDB
3UVJ
E-value=7.60608e-66,
Score=635
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cytoplasm
SignalP
Position: 1 - 17,
Likelihood: 0.590325
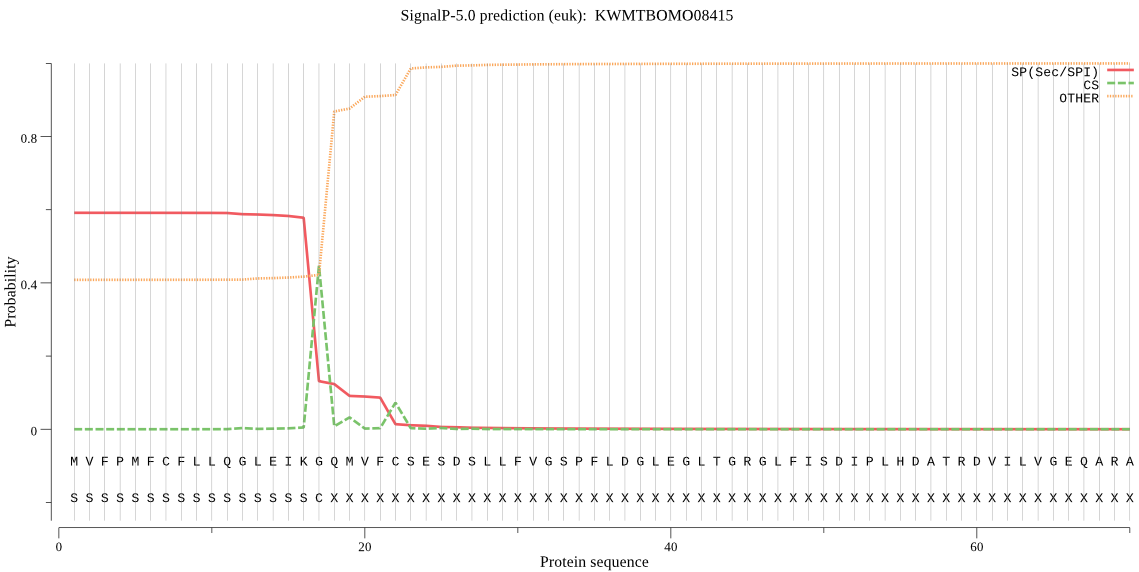
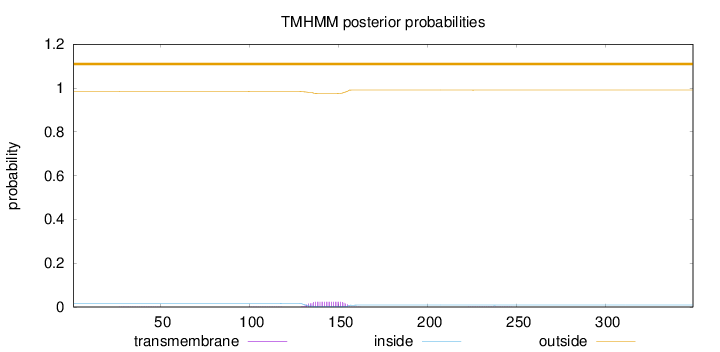
Length:
349
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.55729
Exp number, first 60 AAs:
0.01429
Total prob of N-in:
0.01574
outside
1 - 349
Population Genetic Test Statistics
Pi
253.936441
Theta
204.881293
Tajima's D
1.665669
CLR
0.314106
CSRT
0.825958702064897
Interpretation
Possibly Positive selection
