Gene
KWMTBOMO08396 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009209
Annotation
uncharacterized_protein_LOC101744449_[Bombyx_mori]
Full name
Protein tumorous imaginal discs, mitochondrial
Alternative Name
Protein lethal(2)tumorous imaginal discs
TID58
TID58
Location in the cell
Mitochondrial Reliability : 1.182 Nuclear Reliability : 1.533
Sequence
CDS
ATGGCTCCTGCTCGGAGTGCTCTGGGATTTTTAAATTCAAGAACTTTTAATTCATTATTAACTAATACAATACCTAAAACTACTTATAGATTTTTACATAAATGTAATAATTGTGAACGTTTTGTTGGAAGCCACGCCATATCGGATACTTCTTTCAAAACTTATAATTTCCCGAAACATCGACATATTCACACTACAAATAATCTATATAACCGCGCCGATTATTATAAAGTACTCGGTGTATCCAAAAATGCATCGGCCAAAGACATCAAGAAGGCATACTATCAACTTGCGAAGAAATATCACCCTGACGCCAATAAATCGGACCCAGAAGCTTCGAAAAAGTTTCAAGAAGTATCTGAAGCCTATGAGATATTATCAGATGAAAATAAACGTAAACAATATGACACATATGGCACAACGTCCGAACAAATGGGCATGGGTGGCCCCGGAGGCCCTGATGGTTTCACCCATCAGTGGCAGTACAAATCGACAATAGACCCTGAAGAATTGTTTAGGAAGATTTTTGGAGATGCTGGCTTCAAGAGTAATACTTTTAGTGATTTTGCTGAAAGCCAATTTGGGTTTGGCGCAGCTCAAGAGATTATAGTAAATTTGAGGTTCACTGAAGCAGCTCGAGGTGTTAATAAAGATGTCAATGTTAATGTTGTTGACACCTGTCCAAAATGCCAAGGTTCTCGTTGTGAGCTAGGAACAAAAGCTATAAGATGCACATATTGTAATGGAACTGGAATGGAAACATTTTCAAGAGGTCCATTTGTAATGCGCTCAACATGCAGACATTGTCATGGTACAAGAATGTTGATAAAATACCCGTGTGTTGAATGTGAAGGAAAGGGACAGTCTGTACAACGTAAGAAAATAACAATACCTGTGCCAGCTGGTGTGGAAGATGGTCAGACAGTGCGTATGTCGGTTGGAAACAATGAATTATTTATCACATTTAAAGTGGAGTCTTCGAATTATTTCAAAAGAGACGGTCCTGATGTACATACTGATTGTACAATATCAATTGCTCAAGCTGTGCTTGGTGGTACAGTCAGGATACAGGGACTGTATGAAGATCACACATTACAGATAGTACCGGGAACATCATCTCACAGCACTATTCGTTTATCAAGGAAAGGAATGAAACGTGTCAGCCAACATGGTCACGGTGATCACTACGTACACATTAAAATACAGGTACCAAAGACACTGACAGACAAGCAGAAAGCCTTGATGTATGCTTATGCCGAACTCGAAGAAGACACTCCCGGTCAAATACTAGGAGTTTCCTATGACAGAGATGAAAAAACAAATGATAGTGCTCGGGAAAACGAAAAGATTCACCGTATGAATCGAGATAACGACCAGGGTAATGACAATAGCAGAGGCTGGACATTTGTAGACAGTATCATTGAAACGTTTCGCGAGAACAAACCGGCATTCTTAACTGGTTTCGTAACTGCTGTCATATTGTGTTACTATACGTATTACCAACCGTACTCATACGGCACTACGGAACCGGAAAGGGAGTTAGACCTGCCCATGGGCTACGAGAGAACTATTAAAAAACAAAAGGAGATGCGACCTGATTTGTTCGAAGCGTGA
Protein
MAPARSALGFLNSRTFNSLLTNTIPKTTYRFLHKCNNCERFVGSHAISDTSFKTYNFPKHRHIHTTNNLYNRADYYKVLGVSKNASAKDIKKAYYQLAKKYHPDANKSDPEASKKFQEVSEAYEILSDENKRKQYDTYGTTSEQMGMGGPGGPDGFTHQWQYKSTIDPEELFRKIFGDAGFKSNTFSDFAESQFGFGAAQEIIVNLRFTEAARGVNKDVNVNVVDTCPKCQGSRCELGTKAIRCTYCNGTGMETFSRGPFVMRSTCRHCHGTRMLIKYPCVECEGKGQSVQRKKITIPVPAGVEDGQTVRMSVGNNELFITFKVESSNYFKRDGPDVHTDCTISIAQAVLGGTVRIQGLYEDHTLQIVPGTSSHSTIRLSRKGMKRVSQHGHGDHYVHIKIQVPKTLTDKQKALMYAYAELEEDTPGQILGVSYDRDEKTNDSARENEKIHRMNRDNDQGNDNSRGWTFVDSIIETFRENKPAFLTGFVTAVILCYYTYYQPYSYGTTEPERELDLPMGYERTIKKQKEMRPDLFEA
Summary
Description
May act as a tumor suppressor in larval imaginal disks.
Keywords
Chaperone
Developmental protein
Membrane
Metal-binding
Mitochondrion
Mitochondrion outer membrane
Repeat
Transit peptide
Zinc
Zinc-finger
Feature
chain Protein tumorous imaginal discs, mitochondrial
Uniprot
H6VTQ4
A0A2H1WR11
A0A0N1PF48
A0A2A4IW77
A0A0N1PJQ2
A0A2A4IWW1
+ More
A0A3S2M082 A0A087ZNL6 A0A195B0C2 A0A1J1IXS5 A0A310SNY4 A0A158NR22 A0A195EUA5 A0A0L7R4P0 F4WP97 A0A067QSP8 A0A026WNX8 A0A2J7R8X2 A0A2A3E288 A0A2J7R8W2 A0A151I983 A0A195E5W8 A0A151WMB9 E9J573 D1ZZJ9 A0A232EYQ4 E2BN18 K7ISA8 E2ADT8 Q16R72 A0A1Y1KAV3 A0A1Y1KAZ9 A0A1W4XJ54 A0A1Y1KDI8 A0A182G403 A0A182KCS0 A0A1B6M6S0 A0A1Q3G2L1 A0A1Q3G2A7 A0A1Q3FX59 A0A182IRA8 A0A336LKQ2 A0A1Q3FXA3 A0A182XZ32 A0A1B6MAG6 A0A182P3R3 A0A182KRL4 A0A182WIP0 A0A1Q3FWS5 A0A0C9R9U7 N6UCG7 A0A182X3I7 A0A1Q3FX17 A0A0A1WXC7 A0A0A1WI88 A0A182NH83 A0A1B6LEQ5 A0A182UFB6 A0A182Q0W7 A0A0M8ZSL8 W8BSL0 E0VXY1 W8CDC0 W5JTS9 A0A182UMS4 A0A0C9Q6I2 A0A1I8P162 A0A2M3ZAG6 A0A1I8P1A2 A0A2M4AFS7 A0A1L8E082 A0A034WQ33 A0A0Q9W0F8 A0A034WTQ0 A0A1B6GE62 A0A2M4BME6 W5JHS9 A0A1B6GHS9 A0A182MX04 B4MDV9 A0A2M4CIG3 A0A182FI81 A0A0K8UGV0 J3JWQ6 A0A0K8TY22 A0A182I3I4 T1PAG9 A0A0Q9WDQ9 Q24331 A0A224XF90 A0A2M4BK61 A0A2M4CYY1 A0A1I8MU35 A0A0Q9W9D4 A0A2M4AF25 A0A1L8DZZ3 A0A2M4D0C9
A0A3S2M082 A0A087ZNL6 A0A195B0C2 A0A1J1IXS5 A0A310SNY4 A0A158NR22 A0A195EUA5 A0A0L7R4P0 F4WP97 A0A067QSP8 A0A026WNX8 A0A2J7R8X2 A0A2A3E288 A0A2J7R8W2 A0A151I983 A0A195E5W8 A0A151WMB9 E9J573 D1ZZJ9 A0A232EYQ4 E2BN18 K7ISA8 E2ADT8 Q16R72 A0A1Y1KAV3 A0A1Y1KAZ9 A0A1W4XJ54 A0A1Y1KDI8 A0A182G403 A0A182KCS0 A0A1B6M6S0 A0A1Q3G2L1 A0A1Q3G2A7 A0A1Q3FX59 A0A182IRA8 A0A336LKQ2 A0A1Q3FXA3 A0A182XZ32 A0A1B6MAG6 A0A182P3R3 A0A182KRL4 A0A182WIP0 A0A1Q3FWS5 A0A0C9R9U7 N6UCG7 A0A182X3I7 A0A1Q3FX17 A0A0A1WXC7 A0A0A1WI88 A0A182NH83 A0A1B6LEQ5 A0A182UFB6 A0A182Q0W7 A0A0M8ZSL8 W8BSL0 E0VXY1 W8CDC0 W5JTS9 A0A182UMS4 A0A0C9Q6I2 A0A1I8P162 A0A2M3ZAG6 A0A1I8P1A2 A0A2M4AFS7 A0A1L8E082 A0A034WQ33 A0A0Q9W0F8 A0A034WTQ0 A0A1B6GE62 A0A2M4BME6 W5JHS9 A0A1B6GHS9 A0A182MX04 B4MDV9 A0A2M4CIG3 A0A182FI81 A0A0K8UGV0 J3JWQ6 A0A0K8TY22 A0A182I3I4 T1PAG9 A0A0Q9WDQ9 Q24331 A0A224XF90 A0A2M4BK61 A0A2M4CYY1 A0A1I8MU35 A0A0Q9W9D4 A0A2M4AF25 A0A1L8DZZ3 A0A2M4D0C9
Pubmed
EMBL
JN872889
AFC01228.1
ODYU01010413
SOQ55499.1
KQ459256
KPJ02093.1
+ More
NWSH01005826 PCG63909.1 KQ459986 KPJ18863.1 PCG63908.1 RSAL01000089 RVE48124.1 KQ976691 KYM77906.1 CVRI01000059 CRL03361.1 KQ760662 OAD59526.1 ADTU01023748 KQ981965 KYN31828.1 KQ414657 KOC65823.1 GL888243 EGI64078.1 KK852983 KDR12875.1 KK107139 QOIP01000003 EZA57720.1 RLU24341.1 NEVH01006721 PNF37272.1 KZ288468 PBC25376.1 PNF37273.1 KQ978296 KYM95368.1 KQ979608 KYN20481.1 KQ982944 KYQ49019.1 GL768124 EFZ12024.1 KQ971338 EFA02382.2 NNAY01001615 OXU23417.1 GL449382 EFN82922.1 GL438820 EFN68450.1 CH477722 EAT36907.1 GEZM01087619 GEZM01087618 JAV58569.1 GEZM01087617 JAV58574.1 GEZM01087620 GEZM01087616 JAV58568.1 JXUM01142693 KQ569461 KXJ68621.1 GEBQ01020191 GEBQ01008386 JAT19786.1 JAT31591.1 GFDL01001066 JAV33979.1 GFDL01001111 JAV33934.1 GFDL01002917 JAV32128.1 UFQT01000040 SSX18612.1 GFDL01002867 JAV32178.1 GEBQ01007069 JAT32908.1 GFDL01002985 JAV32060.1 GBYB01009712 JAG79479.1 APGK01029086 KB740724 ENN79355.1 GFDL01002884 JAV32161.1 GBXI01010583 JAD03709.1 GBXI01016179 JAC98112.1 GEBQ01017806 JAT22171.1 AXCN02000893 KQ435865 KOX70310.1 GAMC01002210 JAC04346.1 DS235842 EEB18237.1 GAMC01002211 JAC04345.1 ADMH02000120 ETN67767.1 GBYB01009713 JAG79480.1 GGFM01004762 MBW25513.1 GGFK01006167 MBW39488.1 GFDF01002015 JAV12069.1 GAKP01001271 JAC57681.1 CH940662 KRF78434.1 GAKP01001270 JAC57682.1 GECZ01009069 JAS60700.1 GGFJ01005023 MBW54164.1 ADMH02001474 ETN62455.1 GECZ01007767 JAS62002.1 AXCM01007945 EDW58724.1 GGFL01000520 MBW64698.1 GDHF01026749 JAI25565.1 BT127674 AEE62636.1 GDHF01033138 JAI19176.1 APCN01000213 KA645150 AFP59779.1 KRF78432.1 Y07700 GFTR01006732 JAW09694.1 GGFJ01004037 MBW53178.1 GGFL01006311 MBW70489.1 KRF78433.1 GGFK01006059 MBW39380.1 GFDF01002021 JAV12063.1 GGFL01006380 MBW70558.1
NWSH01005826 PCG63909.1 KQ459986 KPJ18863.1 PCG63908.1 RSAL01000089 RVE48124.1 KQ976691 KYM77906.1 CVRI01000059 CRL03361.1 KQ760662 OAD59526.1 ADTU01023748 KQ981965 KYN31828.1 KQ414657 KOC65823.1 GL888243 EGI64078.1 KK852983 KDR12875.1 KK107139 QOIP01000003 EZA57720.1 RLU24341.1 NEVH01006721 PNF37272.1 KZ288468 PBC25376.1 PNF37273.1 KQ978296 KYM95368.1 KQ979608 KYN20481.1 KQ982944 KYQ49019.1 GL768124 EFZ12024.1 KQ971338 EFA02382.2 NNAY01001615 OXU23417.1 GL449382 EFN82922.1 GL438820 EFN68450.1 CH477722 EAT36907.1 GEZM01087619 GEZM01087618 JAV58569.1 GEZM01087617 JAV58574.1 GEZM01087620 GEZM01087616 JAV58568.1 JXUM01142693 KQ569461 KXJ68621.1 GEBQ01020191 GEBQ01008386 JAT19786.1 JAT31591.1 GFDL01001066 JAV33979.1 GFDL01001111 JAV33934.1 GFDL01002917 JAV32128.1 UFQT01000040 SSX18612.1 GFDL01002867 JAV32178.1 GEBQ01007069 JAT32908.1 GFDL01002985 JAV32060.1 GBYB01009712 JAG79479.1 APGK01029086 KB740724 ENN79355.1 GFDL01002884 JAV32161.1 GBXI01010583 JAD03709.1 GBXI01016179 JAC98112.1 GEBQ01017806 JAT22171.1 AXCN02000893 KQ435865 KOX70310.1 GAMC01002210 JAC04346.1 DS235842 EEB18237.1 GAMC01002211 JAC04345.1 ADMH02000120 ETN67767.1 GBYB01009713 JAG79480.1 GGFM01004762 MBW25513.1 GGFK01006167 MBW39488.1 GFDF01002015 JAV12069.1 GAKP01001271 JAC57681.1 CH940662 KRF78434.1 GAKP01001270 JAC57682.1 GECZ01009069 JAS60700.1 GGFJ01005023 MBW54164.1 ADMH02001474 ETN62455.1 GECZ01007767 JAS62002.1 AXCM01007945 EDW58724.1 GGFL01000520 MBW64698.1 GDHF01026749 JAI25565.1 BT127674 AEE62636.1 GDHF01033138 JAI19176.1 APCN01000213 KA645150 AFP59779.1 KRF78432.1 Y07700 GFTR01006732 JAW09694.1 GGFJ01004037 MBW53178.1 GGFL01006311 MBW70489.1 KRF78433.1 GGFK01006059 MBW39380.1 GFDF01002021 JAV12063.1 GGFL01006380 MBW70558.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000283053
UP000005203
UP000078540
+ More
UP000183832 UP000005205 UP000078541 UP000053825 UP000007755 UP000027135 UP000053097 UP000279307 UP000235965 UP000242457 UP000078542 UP000078492 UP000075809 UP000007266 UP000215335 UP000008237 UP000002358 UP000000311 UP000008820 UP000192223 UP000069940 UP000249989 UP000075881 UP000075880 UP000076408 UP000075885 UP000075882 UP000075920 UP000019118 UP000076407 UP000075884 UP000075902 UP000075886 UP000053105 UP000009046 UP000000673 UP000075903 UP000095300 UP000008792 UP000075883 UP000069272 UP000075840 UP000095301
UP000183832 UP000005205 UP000078541 UP000053825 UP000007755 UP000027135 UP000053097 UP000279307 UP000235965 UP000242457 UP000078542 UP000078492 UP000075809 UP000007266 UP000215335 UP000008237 UP000002358 UP000000311 UP000008820 UP000192223 UP000069940 UP000249989 UP000075881 UP000075880 UP000076408 UP000075885 UP000075882 UP000075920 UP000019118 UP000076407 UP000075884 UP000075902 UP000075886 UP000053105 UP000009046 UP000000673 UP000075903 UP000095300 UP000008792 UP000075883 UP000069272 UP000075840 UP000095301
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H6VTQ4
A0A2H1WR11
A0A0N1PF48
A0A2A4IW77
A0A0N1PJQ2
A0A2A4IWW1
+ More
A0A3S2M082 A0A087ZNL6 A0A195B0C2 A0A1J1IXS5 A0A310SNY4 A0A158NR22 A0A195EUA5 A0A0L7R4P0 F4WP97 A0A067QSP8 A0A026WNX8 A0A2J7R8X2 A0A2A3E288 A0A2J7R8W2 A0A151I983 A0A195E5W8 A0A151WMB9 E9J573 D1ZZJ9 A0A232EYQ4 E2BN18 K7ISA8 E2ADT8 Q16R72 A0A1Y1KAV3 A0A1Y1KAZ9 A0A1W4XJ54 A0A1Y1KDI8 A0A182G403 A0A182KCS0 A0A1B6M6S0 A0A1Q3G2L1 A0A1Q3G2A7 A0A1Q3FX59 A0A182IRA8 A0A336LKQ2 A0A1Q3FXA3 A0A182XZ32 A0A1B6MAG6 A0A182P3R3 A0A182KRL4 A0A182WIP0 A0A1Q3FWS5 A0A0C9R9U7 N6UCG7 A0A182X3I7 A0A1Q3FX17 A0A0A1WXC7 A0A0A1WI88 A0A182NH83 A0A1B6LEQ5 A0A182UFB6 A0A182Q0W7 A0A0M8ZSL8 W8BSL0 E0VXY1 W8CDC0 W5JTS9 A0A182UMS4 A0A0C9Q6I2 A0A1I8P162 A0A2M3ZAG6 A0A1I8P1A2 A0A2M4AFS7 A0A1L8E082 A0A034WQ33 A0A0Q9W0F8 A0A034WTQ0 A0A1B6GE62 A0A2M4BME6 W5JHS9 A0A1B6GHS9 A0A182MX04 B4MDV9 A0A2M4CIG3 A0A182FI81 A0A0K8UGV0 J3JWQ6 A0A0K8TY22 A0A182I3I4 T1PAG9 A0A0Q9WDQ9 Q24331 A0A224XF90 A0A2M4BK61 A0A2M4CYY1 A0A1I8MU35 A0A0Q9W9D4 A0A2M4AF25 A0A1L8DZZ3 A0A2M4D0C9
A0A3S2M082 A0A087ZNL6 A0A195B0C2 A0A1J1IXS5 A0A310SNY4 A0A158NR22 A0A195EUA5 A0A0L7R4P0 F4WP97 A0A067QSP8 A0A026WNX8 A0A2J7R8X2 A0A2A3E288 A0A2J7R8W2 A0A151I983 A0A195E5W8 A0A151WMB9 E9J573 D1ZZJ9 A0A232EYQ4 E2BN18 K7ISA8 E2ADT8 Q16R72 A0A1Y1KAV3 A0A1Y1KAZ9 A0A1W4XJ54 A0A1Y1KDI8 A0A182G403 A0A182KCS0 A0A1B6M6S0 A0A1Q3G2L1 A0A1Q3G2A7 A0A1Q3FX59 A0A182IRA8 A0A336LKQ2 A0A1Q3FXA3 A0A182XZ32 A0A1B6MAG6 A0A182P3R3 A0A182KRL4 A0A182WIP0 A0A1Q3FWS5 A0A0C9R9U7 N6UCG7 A0A182X3I7 A0A1Q3FX17 A0A0A1WXC7 A0A0A1WI88 A0A182NH83 A0A1B6LEQ5 A0A182UFB6 A0A182Q0W7 A0A0M8ZSL8 W8BSL0 E0VXY1 W8CDC0 W5JTS9 A0A182UMS4 A0A0C9Q6I2 A0A1I8P162 A0A2M3ZAG6 A0A1I8P1A2 A0A2M4AFS7 A0A1L8E082 A0A034WQ33 A0A0Q9W0F8 A0A034WTQ0 A0A1B6GE62 A0A2M4BME6 W5JHS9 A0A1B6GHS9 A0A182MX04 B4MDV9 A0A2M4CIG3 A0A182FI81 A0A0K8UGV0 J3JWQ6 A0A0K8TY22 A0A182I3I4 T1PAG9 A0A0Q9WDQ9 Q24331 A0A224XF90 A0A2M4BK61 A0A2M4CYY1 A0A1I8MU35 A0A0Q9W9D4 A0A2M4AF25 A0A1L8DZZ3 A0A2M4D0C9
PDB
2DN9
E-value=6.19032e-22,
Score=258
Ontologies
GO
Topology
Subcellular location
Mitochondrion outer membrane
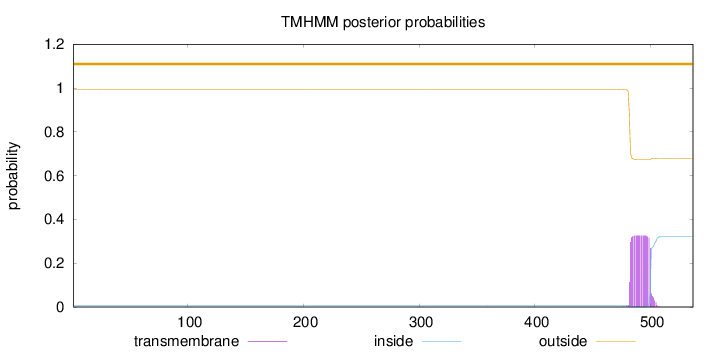
Length:
537
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.10012
Exp number, first 60 AAs:
0.00212
Total prob of N-in:
0.00608
outside
1 - 537
Population Genetic Test Statistics
Pi
276.943642
Theta
192.537047
Tajima's D
1.371149
CLR
0.614331
CSRT
0.75911204439778
Interpretation
Uncertain
