Gene
KWMTBOMO08393
Pre Gene Modal
BGIBMGA014515
Annotation
PREDICTED:_uncharacterized_protein_LOC101736465_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.304
Sequence
CDS
ATGTGCGTGAAAAATGACGAGATGCCTACGGATATTCGGCATCAAGAACCCGAACAGCTGCAGACGTCAATCGGTGATTTTGACTATAAGCCAGCCTCATTCTTGATTGGGATGCAGCCAAACGAAAGCATAAATTACTGCGATTTTAATTACGGCCTCCCGCACACGCTCGAATGGGATATCCTCCAAACTTATCCGTCCCCGGAGACTGGAAATAAAAGTCCTTATAGAGTCATATATGATGCTATCAAAAGTAGGGCTTATAGTAATTGCGGTGTGTATAATGCAGTAACTTATGCCACACAAGCAACTCCTGAATTCATTTACCATATCGTTGAGATAGCTCGCTACTGGGATGGACCCATCAGTCTCAGTGTATTTGTACCTGATTACGATTTGGATATAACGATGCAGATAATGAACCAGCTCTGCCATTGCTACCCTGGTATGTCCAAAGTTTCATTACATCTCTTTTATCCAAAGAAACTCACACCAAAACTTAGAACCAGGAGGCTGTTTTTACCTGTTACTACGGAACCGTCAACGACACCATCGAACATCAGCTTAGAAGAGATGTTCAGGAAGAAATTGGAAAATTACAGGAATTTGAACAACAAAACTAGAGCTGAGTACATACAAGGAGCAAGGAAAAGAAAAATTGAAAAGATGATGATCGGCATGATCAAAGGAAAACCGGTTATACCGAAGTTAACATTCAAAGATTGTTCGGGTCCAGATTCGTTCGATATAAAAACATTTAGAAAGGAAAACAACATGCTATATCCGATTAATGTCGGCAGGAACGTTGCTAGAAATGCATCGAAGACGAACTATTTCATAGTTTCCGATATAGAAATGGTGCCGAGTGACGGGTTAGCGACTAAATTTCTCGTGATGCTGAGGAAACTTATGGGAAATAAGAAACGCGACGAGGGCTGTGTGTTCACTAAGACGATCTTTGTGGTGCCATTGTTTGAAGTGGAGAGAGGCGAGGAAATACCGCGGAACAAAGACACATTAGTTCGCATGGTAGCGGCGAACCGAGCAATGTATTTCCATCAGAAGGTCTGCTCGCATTGTCAAAGGTTTCCCGGACTGCAGTCGTGGCTTCTGCGAACCCCGCCGGACGTTCTGGAACCGATGCTGATCGCGAGAAGAGAGTACCCGTACCACAGATGGGAGCCTTTGTACTTTGGCACACACAAGGAGCCTTGGTACAGCGAGATGCTCTCTTGGGAGGGACGTCAGGATAAAATGCCACAGATGTTAGAGTTATGCCTTCAAGAATACCGAATGGTGGTCCTGGATGGAGGGTTTCTCTGCCACGCTGCGGTTTCCCGAACTCATTCCACCAGACAATCGCGCGCTGAGAGACTAAACTCGGAACGTTACATGAAGATCTTAGCGGCGCTACGACGTAAATACCATCAGAAACAACAGTGCAAAGTTATGTGA
Protein
MCVKNDEMPTDIRHQEPEQLQTSIGDFDYKPASFLIGMQPNESINYCDFNYGLPHTLEWDILQTYPSPETGNKSPYRVIYDAIKSRAYSNCGVYNAVTYATQATPEFIYHIVEIARYWDGPISLSVFVPDYDLDITMQIMNQLCHCYPGMSKVSLHLFYPKKLTPKLRTRRLFLPVTTEPSTTPSNISLEEMFRKKLENYRNLNNKTRAEYIQGARKRKIEKMMIGMIKGKPVIPKLTFKDCSGPDSFDIKTFRKENNMLYPINVGRNVARNASKTNYFIVSDIEMVPSDGLATKFLVMLRKLMGNKKRDEGCVFTKTIFVVPLFEVERGEEIPRNKDTLVRMVAANRAMYFHQKVCSHCQRFPGLQSWLLRTPPDVLEPMLIARREYPYHRWEPLYFGTHKEPWYSEMLSWEGRQDKMPQMLELCLQEYRMVVLDGGFLCHAAVSRTHSTRQSRAERLNSERYMKILAALRRKYHQKQQCKVM
Summary
Uniprot
A0A437B744
A0A0N1IA20
A0A2A4IVZ0
A0A0N0PE18
A0A0L7LCU5
A0A2H1WK27
+ More
A0A067QV11 A0A2J7R8U9 E0W463 A0A1B6CXQ4 A0A2R7WGI2 E2ADT6 A0A1B6CCH4 A0A2A3E101 A0A232EHY4 E9J575 A0A423SJB4 A0A131ZWU7 T1I0W7 A0A087UR85 A0A154PF67 A0A443QXL3 A0A0P5ES47 A0A3S4QPI2 A0A164V5W6 A0A0P5FKI5 A0A0P5M038 A0A0P5DSI7 A0A2L2YDJ1 A0A2L2YB94 A0A443SLW2 T1JBZ4 A0A0P6H4Z8 A0A0P6BMR5 A0A0P5R751 T1JJI3 A0A226DCR8 A0A1S3DMS8 A0A131Z0G0 A0A336KW97 A0A0N8CTR4 A0A1S4EG64 L7MAL0 A0A1W4WP15 A0A226EP60 A0A0C9RM78 E9GZC9 A0A1V9XQX1 A0A182QFD2 A0A0P6ER66 A0A0P5LJR5 A0A084VLW4 A0A182VMM1 A0A182VUQ8 B7PHJ7 A0A182NEN0 A0A182LZI6 Q7QGB8 A0A293N8I8 A0A0K8SZ03 A0A2R5LE75 A0A1S4H118 A0A182QU62 A0A1E1WVN5 A0A182HG70 A0A146LP98 V5GVL7 A0A1E1XPP9 A0A224XE45 W5J378 Q5TVI0 U5ES29 A0A1S4H111 A0A182XDK3 A0A182V5M8
A0A067QV11 A0A2J7R8U9 E0W463 A0A1B6CXQ4 A0A2R7WGI2 E2ADT6 A0A1B6CCH4 A0A2A3E101 A0A232EHY4 E9J575 A0A423SJB4 A0A131ZWU7 T1I0W7 A0A087UR85 A0A154PF67 A0A443QXL3 A0A0P5ES47 A0A3S4QPI2 A0A164V5W6 A0A0P5FKI5 A0A0P5M038 A0A0P5DSI7 A0A2L2YDJ1 A0A2L2YB94 A0A443SLW2 T1JBZ4 A0A0P6H4Z8 A0A0P6BMR5 A0A0P5R751 T1JJI3 A0A226DCR8 A0A1S3DMS8 A0A131Z0G0 A0A336KW97 A0A0N8CTR4 A0A1S4EG64 L7MAL0 A0A1W4WP15 A0A226EP60 A0A0C9RM78 E9GZC9 A0A1V9XQX1 A0A182QFD2 A0A0P6ER66 A0A0P5LJR5 A0A084VLW4 A0A182VMM1 A0A182VUQ8 B7PHJ7 A0A182NEN0 A0A182LZI6 Q7QGB8 A0A293N8I8 A0A0K8SZ03 A0A2R5LE75 A0A1S4H118 A0A182QU62 A0A1E1WVN5 A0A182HG70 A0A146LP98 V5GVL7 A0A1E1XPP9 A0A224XE45 W5J378 Q5TVI0 U5ES29 A0A1S4H111 A0A182XDK3 A0A182V5M8
Pubmed
EMBL
RSAL01000133
RVE46337.1
KQ459256
KPJ02094.1
NWSH01005826
PCG63911.1
+ More
KQ459986 KPJ18862.1 JTDY01001629 KOB73307.1 ODYU01009182 SOQ53431.1 KK852983 KDR12877.1 NEVH01006721 PNF37263.1 DS235886 EEB20419.1 GEDC01023767 GEDC01019110 JAS13531.1 JAS18188.1 KK854722 PTY18170.1 GL438820 EFN68448.1 GEDC01026150 JAS11148.1 KZ288468 PBC25378.1 NNAY01004419 OXU17931.1 GL768124 EFZ12031.1 QCYY01003277 ROT64296.1 JXLN01002828 KPM02765.1 ACPB03010575 KK121161 KFM79874.1 KQ434879 KZC09948.1 NCKU01003324 RWS07749.1 GDIQ01265784 JAJ85940.1 NCKU01004341 NCKU01004340 RWS06097.1 RWS06099.1 LRGB01001411 KZS11999.1 GDIQ01261415 JAJ90309.1 GDIQ01171678 JAK80047.1 GDIP01151239 JAJ72163.1 IAAA01010261 LAA05420.1 IAAA01010260 LAA05414.1 NCKV01001328 RWS28526.1 JH432044 GDIQ01024457 JAN70280.1 GDIP01012481 JAM91234.1 GDIQ01104722 JAL47004.1 JH430212 LNIX01000025 OXA42728.1 GEDV01003638 JAP84919.1 UFQS01001083 UFQT01001083 SSX08892.1 SSX28803.1 GDIP01099735 JAM03980.1 GACK01004880 JAA60154.1 LNIX01000003 OXA58356.1 GBYB01006727 GBYB01009395 GBYB01010023 JAG76494.1 JAG79162.1 JAG79790.1 GL732577 EFX75183.1 MNPL01005615 OQR75906.1 AXCN02000847 GDIQ01059006 JAN35731.1 GDIQ01172614 JAK79111.1 ATLV01014570 KE524975 KFB38958.1 ABJB010544036 DS713392 EEC06069.1 AXCM01000061 AAAB01008835 EAA05791.3 GFWV01023243 MAA47970.1 GBRD01007681 JAG58140.1 GGLE01003531 MBY07657.1 GDQN01000203 JAT90851.1 APCN01004181 APCN01004182 APCN01004183 GDHC01008911 JAQ09718.1 GALX01004183 JAB64283.1 GFAA01002124 JAU01311.1 GFTR01005701 JAW10725.1 ADMH02002118 ETN58802.1 EAL41415.3 GANO01003433 JAB56438.1
KQ459986 KPJ18862.1 JTDY01001629 KOB73307.1 ODYU01009182 SOQ53431.1 KK852983 KDR12877.1 NEVH01006721 PNF37263.1 DS235886 EEB20419.1 GEDC01023767 GEDC01019110 JAS13531.1 JAS18188.1 KK854722 PTY18170.1 GL438820 EFN68448.1 GEDC01026150 JAS11148.1 KZ288468 PBC25378.1 NNAY01004419 OXU17931.1 GL768124 EFZ12031.1 QCYY01003277 ROT64296.1 JXLN01002828 KPM02765.1 ACPB03010575 KK121161 KFM79874.1 KQ434879 KZC09948.1 NCKU01003324 RWS07749.1 GDIQ01265784 JAJ85940.1 NCKU01004341 NCKU01004340 RWS06097.1 RWS06099.1 LRGB01001411 KZS11999.1 GDIQ01261415 JAJ90309.1 GDIQ01171678 JAK80047.1 GDIP01151239 JAJ72163.1 IAAA01010261 LAA05420.1 IAAA01010260 LAA05414.1 NCKV01001328 RWS28526.1 JH432044 GDIQ01024457 JAN70280.1 GDIP01012481 JAM91234.1 GDIQ01104722 JAL47004.1 JH430212 LNIX01000025 OXA42728.1 GEDV01003638 JAP84919.1 UFQS01001083 UFQT01001083 SSX08892.1 SSX28803.1 GDIP01099735 JAM03980.1 GACK01004880 JAA60154.1 LNIX01000003 OXA58356.1 GBYB01006727 GBYB01009395 GBYB01010023 JAG76494.1 JAG79162.1 JAG79790.1 GL732577 EFX75183.1 MNPL01005615 OQR75906.1 AXCN02000847 GDIQ01059006 JAN35731.1 GDIQ01172614 JAK79111.1 ATLV01014570 KE524975 KFB38958.1 ABJB010544036 DS713392 EEC06069.1 AXCM01000061 AAAB01008835 EAA05791.3 GFWV01023243 MAA47970.1 GBRD01007681 JAG58140.1 GGLE01003531 MBY07657.1 GDQN01000203 JAT90851.1 APCN01004181 APCN01004182 APCN01004183 GDHC01008911 JAQ09718.1 GALX01004183 JAB64283.1 GFAA01002124 JAU01311.1 GFTR01005701 JAW10725.1 ADMH02002118 ETN58802.1 EAL41415.3 GANO01003433 JAB56438.1
Proteomes
UP000283053
UP000053268
UP000218220
UP000053240
UP000037510
UP000027135
+ More
UP000235965 UP000009046 UP000000311 UP000242457 UP000215335 UP000283509 UP000015103 UP000054359 UP000076502 UP000285301 UP000076858 UP000288716 UP000198287 UP000079169 UP000192223 UP000000305 UP000192247 UP000075886 UP000030765 UP000075903 UP000075920 UP000001555 UP000075884 UP000075883 UP000007062 UP000075840 UP000000673 UP000076407
UP000235965 UP000009046 UP000000311 UP000242457 UP000215335 UP000283509 UP000015103 UP000054359 UP000076502 UP000285301 UP000076858 UP000288716 UP000198287 UP000079169 UP000192223 UP000000305 UP000192247 UP000075886 UP000030765 UP000075903 UP000075920 UP000001555 UP000075884 UP000075883 UP000007062 UP000075840 UP000000673 UP000076407
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
A0A437B744
A0A0N1IA20
A0A2A4IVZ0
A0A0N0PE18
A0A0L7LCU5
A0A2H1WK27
+ More
A0A067QV11 A0A2J7R8U9 E0W463 A0A1B6CXQ4 A0A2R7WGI2 E2ADT6 A0A1B6CCH4 A0A2A3E101 A0A232EHY4 E9J575 A0A423SJB4 A0A131ZWU7 T1I0W7 A0A087UR85 A0A154PF67 A0A443QXL3 A0A0P5ES47 A0A3S4QPI2 A0A164V5W6 A0A0P5FKI5 A0A0P5M038 A0A0P5DSI7 A0A2L2YDJ1 A0A2L2YB94 A0A443SLW2 T1JBZ4 A0A0P6H4Z8 A0A0P6BMR5 A0A0P5R751 T1JJI3 A0A226DCR8 A0A1S3DMS8 A0A131Z0G0 A0A336KW97 A0A0N8CTR4 A0A1S4EG64 L7MAL0 A0A1W4WP15 A0A226EP60 A0A0C9RM78 E9GZC9 A0A1V9XQX1 A0A182QFD2 A0A0P6ER66 A0A0P5LJR5 A0A084VLW4 A0A182VMM1 A0A182VUQ8 B7PHJ7 A0A182NEN0 A0A182LZI6 Q7QGB8 A0A293N8I8 A0A0K8SZ03 A0A2R5LE75 A0A1S4H118 A0A182QU62 A0A1E1WVN5 A0A182HG70 A0A146LP98 V5GVL7 A0A1E1XPP9 A0A224XE45 W5J378 Q5TVI0 U5ES29 A0A1S4H111 A0A182XDK3 A0A182V5M8
A0A067QV11 A0A2J7R8U9 E0W463 A0A1B6CXQ4 A0A2R7WGI2 E2ADT6 A0A1B6CCH4 A0A2A3E101 A0A232EHY4 E9J575 A0A423SJB4 A0A131ZWU7 T1I0W7 A0A087UR85 A0A154PF67 A0A443QXL3 A0A0P5ES47 A0A3S4QPI2 A0A164V5W6 A0A0P5FKI5 A0A0P5M038 A0A0P5DSI7 A0A2L2YDJ1 A0A2L2YB94 A0A443SLW2 T1JBZ4 A0A0P6H4Z8 A0A0P6BMR5 A0A0P5R751 T1JJI3 A0A226DCR8 A0A1S3DMS8 A0A131Z0G0 A0A336KW97 A0A0N8CTR4 A0A1S4EG64 L7MAL0 A0A1W4WP15 A0A226EP60 A0A0C9RM78 E9GZC9 A0A1V9XQX1 A0A182QFD2 A0A0P6ER66 A0A0P5LJR5 A0A084VLW4 A0A182VMM1 A0A182VUQ8 B7PHJ7 A0A182NEN0 A0A182LZI6 Q7QGB8 A0A293N8I8 A0A0K8SZ03 A0A2R5LE75 A0A1S4H118 A0A182QU62 A0A1E1WVN5 A0A182HG70 A0A146LP98 V5GVL7 A0A1E1XPP9 A0A224XE45 W5J378 Q5TVI0 U5ES29 A0A1S4H111 A0A182XDK3 A0A182V5M8
Ontologies
PATHWAY
GO
PANTHER
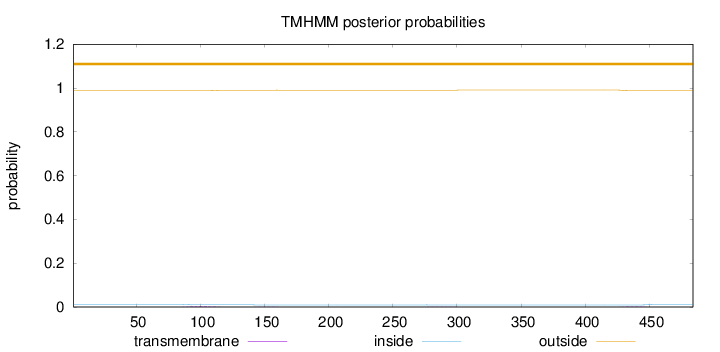
Topology
Length:
484
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10251
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01256
outside
1 - 484
Population Genetic Test Statistics
Pi
212.456802
Theta
173.520088
Tajima's D
1.18813
CLR
0.594047
CSRT
0.71666416679166
Interpretation
Uncertain
