Gene
KWMTBOMO08386
Pre Gene Modal
BGIBMGA009212
Annotation
PREDICTED:_uncharacterized_protein_LOC101744019_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.611
Sequence
CDS
ATGGCCAGTGGAAAGATACTGTATTCGATAGCGGTAGTTGTACTCGGCTTCATATGCTTTGTTGTCGGAGCTGTTGCTGTAGGGCTACCTGCTTGGGGATATTTCTTTAGTTATGATTCTGACCAACGATACTCTCCACAGTCTTATTTTTTGGAGAATCCAAACTTCGACCAAGGCTACTTCGGACCATGGCGTGTTTGCAAGAAGTTGCTCTACAACAGGGAGAAATGTGGGACGGACGTTTCGAAGTTCCGACCGTCAACTGCCGTTTACATTTCGGGAGTGGTCGCAGTAGTAGGCGTGACGATCCTGGGGATCTTCTGCATACTGTCAGTGCTTCAGCTAGCGATGATCTCGTCAAAAGAACGTGTCGCCATGAAGTACACGCACCTGGTTATGATGAAGCTGTCTTTAGCTCTGTTGGCAACATTACTGTCAATAGCGGCAGCCGGTCTGTTCGCCGTCCAAGGCGATGACAGGGGTTTCTATTTGAAGAGAGGAGAGGCGTTTTACGTCCAGATAGTATGCATAGTAATTAATTCAGTGCTGTTCATAATGTCGTTGTACGACACCCTGTTCTCACGAAGAGTTGGAGGAGATCCCACGAATCCCCTGGGCGAACCTGACAGTACGACCATCAACAACCCCGGCTTCAAAGATGGAAGAGGGATATCGATGACGGACGCTTCCGGTAAGCCGTACAGCACCAACGGCCACGGCAGCGTCGCCTCCATGAACACAACCGCCACAACGCTTACGGGGCTGTCGGATGCCAGCACCATCACCAGGTCGCCGCTGCGGTCCTCGCTGAAGAAGCCGCGACCAAGAGAAGGGGAACCGGGCGGCCTGGGTATACAGAACCCAGGGTACTCGGGACATTCGCCGCCCTTAAACAGGAACGGAAGCCAGAAGAAAGTGCGCATCCAGACTCACAGCACCGAGGTATGA
Protein
MASGKILYSIAVVVLGFICFVVGAVAVGLPAWGYFFSYDSDQRYSPQSYFLENPNFDQGYFGPWRVCKKLLYNREKCGTDVSKFRPSTAVYISGVVAVVGVTILGIFCILSVLQLAMISSKERVAMKYTHLVMMKLSLALLATLLSIAAAGLFAVQGDDRGFYLKRGEAFYVQIVCIVINSVLFIMSLYDTLFSRRVGGDPTNPLGEPDSTTINNPGFKDGRGISMTDASGKPYSTNGHGSVASMNTTATTLTGLSDASTITRSPLRSSLKKPRPREGEPGGLGIQNPGYSGHSPPLNRNGSQKKVRIQTHSTEV
Summary
Uniprot
A0A2W1B457
A0A212FK78
A0A0N1IE54
A0A1I8MDT8
A0A1Y1LP15
U4URT7
+ More
A0A0L0CKB6 A0A1I8NWS6 D2A1J6 A0A1A9X0Z5 B4QPE3 A0A1W4VS70 Q9VVN1 B4IFP8 Q2LZ07 B4PJF2 A0A0P8ZTY6 B3ND98 B4LF71 B3M3K4 A0A1B0CAN6 W8C5D0 A0A3B0J5F8 U5EPU8 B4J3A4 A0A0M3QX50 B4MLG6 A0A1B0DJ96 A0A1L8D9Y5 A0A0K8UBW9 A0A034V6W1 A0A069DSM9 A0A146KVN9 A0A1B6DRN9 A0A0A9W3R8 A0A1A9UM44 A0A1A9ZDL1 A0A1B6HHV3 A0A2S2RB87 A0A1B6G8W7 A0A1J1HY90 X1WJ41 A0A2S2PA68 B4GZU0 A0A2M3ZEK6 W5JT44 A0A2M4BUD3 A0A1Q3FM42 A0A336LYS8 A0A1Q3FMN7 A0A2M4A566 A0A336KY36 A0A1Q3FM64 A0A182FHN0 A0A023EQP5 A0A023END2 A0A182GJY6 A0A1A9Y1N4 A0A182NMB8 A0A182RC11 K7ISB0 A0A182WLV2 A0A182QDN6 A0A182JJ00 A0A2A3ESF2 Q7Q790 A0A026WQC0 A0A182K069 A0A182URG7 A0A182HM76 A0A182MUD6 B0X3D2 A0A084VD88 A0A023ENU8 B0WT66 A0A195E5Z9 B0X3D1 A0A151IH69 A0A088AGX2 A0A195F3A1 A0A195BAC1 A0A151X6E0 A0A182T0Y4 T1GF29 E0VCG9 A0A154P8U3 A0A182PVV0
A0A0L0CKB6 A0A1I8NWS6 D2A1J6 A0A1A9X0Z5 B4QPE3 A0A1W4VS70 Q9VVN1 B4IFP8 Q2LZ07 B4PJF2 A0A0P8ZTY6 B3ND98 B4LF71 B3M3K4 A0A1B0CAN6 W8C5D0 A0A3B0J5F8 U5EPU8 B4J3A4 A0A0M3QX50 B4MLG6 A0A1B0DJ96 A0A1L8D9Y5 A0A0K8UBW9 A0A034V6W1 A0A069DSM9 A0A146KVN9 A0A1B6DRN9 A0A0A9W3R8 A0A1A9UM44 A0A1A9ZDL1 A0A1B6HHV3 A0A2S2RB87 A0A1B6G8W7 A0A1J1HY90 X1WJ41 A0A2S2PA68 B4GZU0 A0A2M3ZEK6 W5JT44 A0A2M4BUD3 A0A1Q3FM42 A0A336LYS8 A0A1Q3FMN7 A0A2M4A566 A0A336KY36 A0A1Q3FM64 A0A182FHN0 A0A023EQP5 A0A023END2 A0A182GJY6 A0A1A9Y1N4 A0A182NMB8 A0A182RC11 K7ISB0 A0A182WLV2 A0A182QDN6 A0A182JJ00 A0A2A3ESF2 Q7Q790 A0A026WQC0 A0A182K069 A0A182URG7 A0A182HM76 A0A182MUD6 B0X3D2 A0A084VD88 A0A023ENU8 B0WT66 A0A195E5Z9 B0X3D1 A0A151IH69 A0A088AGX2 A0A195F3A1 A0A195BAC1 A0A151X6E0 A0A182T0Y4 T1GF29 E0VCG9 A0A154P8U3 A0A182PVV0
Pubmed
28756777
22118469
26354079
25315136
28004739
23537049
+ More
26108605 18362917 19820115 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 17550304 18057021 24495485 25348373 26334808 26823975 25401762 20920257 23761445 24945155 26483478 20075255 12364791 14747013 17210077 24508170 30249741 24438588 20566863
26108605 18362917 19820115 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 17550304 18057021 24495485 25348373 26334808 26823975 25401762 20920257 23761445 24945155 26483478 20075255 12364791 14747013 17210077 24508170 30249741 24438588 20566863
EMBL
KZ150354
PZC71242.1
AGBW02008113
OWR54148.1
KQ458751
KPJ05323.1
+ More
GEZM01053345 JAV74080.1 KB632337 ERL92846.1 JRES01000296 KNC32662.1 KQ971338 EFA01532.2 CM000363 CM002912 EDX10937.1 KMZ00364.1 AE014296 AY051547 AAF49280.1 AAK92971.1 CH480834 EDW46485.1 CH379069 EAL29702.1 KRT08297.1 CM000159 EDW94640.1 KRK01960.1 CH902618 KPU77978.1 KPU77979.1 CH954178 EDV51891.1 KQS44001.1 CH940647 EDW70259.1 KRF84811.1 KRF84812.1 KRF84813.1 EDV39265.1 AJWK01004130 AJWK01004131 AJWK01004132 GAMC01001833 GAMC01001832 GAMC01001831 GAMC01001830 JAC04724.1 OUUW01000002 SPP77037.1 GANO01004607 JAB55264.1 CH916366 EDV97203.1 CP012525 ALC43516.1 ALC45249.1 CH963847 EDW72822.1 AJVK01034910 AJVK01034911 GFDF01010909 JAV03175.1 GDHF01028231 JAI24083.1 GAKP01020726 GAKP01020723 GAKP01020722 GAKP01020721 JAC38229.1 GBGD01002207 JAC86682.1 GDHC01019452 JAP99176.1 GEDC01012420 GEDC01008963 JAS24878.1 JAS28335.1 GBHO01041548 JAG02056.1 GECU01033515 GECU01003090 JAS74191.1 JAT04617.1 GGMS01017419 MBY86622.1 GECZ01024299 GECZ01010871 GECZ01008259 GECZ01004068 JAS45470.1 JAS58898.1 JAS61510.1 JAS65701.1 CVRI01000036 CRK92989.1 ABLF02039643 ABLF02039644 ABLF02057225 GGMR01013207 MBY25826.1 CH479199 EDW29517.1 GGFM01006109 MBW26860.1 ADMH02000523 ETN66115.1 GGFJ01007534 MBW56675.1 GFDL01006428 JAV28617.1 UFQS01000322 UFQT01000322 SSX02869.1 SSX23236.1 GFDL01006299 JAV28746.1 GGFK01002467 MBW35788.1 UFQS01001198 UFQT01001198 SSX09651.1 SSX29447.1 GFDL01006431 JAV28614.1 GAPW01002669 JAC10929.1 GAPW01002670 JAC10928.1 JXUM01068902 KQ562524 KXJ75702.1 AXCN02000935 KZ288189 PBC34658.1 AAAB01008960 EAA11146.3 KK107139 QOIP01000003 EZA57891.1 RLU24375.1 APCN01004377 AXCM01004374 DS232312 EDS39824.1 ATLV01011192 KE524650 KFB35932.1 GAPW01002983 JAC10615.1 DS232080 EDS34209.1 KQ979608 KYN20279.1 EDS39823.1 KQ977643 KYN00870.1 KQ981836 KYN35055.1 KQ976542 KYM81139.1 KQ982482 KYQ55848.1 CAQQ02092546 DS235053 EEB11075.1 KQ434827 KZC07560.1
GEZM01053345 JAV74080.1 KB632337 ERL92846.1 JRES01000296 KNC32662.1 KQ971338 EFA01532.2 CM000363 CM002912 EDX10937.1 KMZ00364.1 AE014296 AY051547 AAF49280.1 AAK92971.1 CH480834 EDW46485.1 CH379069 EAL29702.1 KRT08297.1 CM000159 EDW94640.1 KRK01960.1 CH902618 KPU77978.1 KPU77979.1 CH954178 EDV51891.1 KQS44001.1 CH940647 EDW70259.1 KRF84811.1 KRF84812.1 KRF84813.1 EDV39265.1 AJWK01004130 AJWK01004131 AJWK01004132 GAMC01001833 GAMC01001832 GAMC01001831 GAMC01001830 JAC04724.1 OUUW01000002 SPP77037.1 GANO01004607 JAB55264.1 CH916366 EDV97203.1 CP012525 ALC43516.1 ALC45249.1 CH963847 EDW72822.1 AJVK01034910 AJVK01034911 GFDF01010909 JAV03175.1 GDHF01028231 JAI24083.1 GAKP01020726 GAKP01020723 GAKP01020722 GAKP01020721 JAC38229.1 GBGD01002207 JAC86682.1 GDHC01019452 JAP99176.1 GEDC01012420 GEDC01008963 JAS24878.1 JAS28335.1 GBHO01041548 JAG02056.1 GECU01033515 GECU01003090 JAS74191.1 JAT04617.1 GGMS01017419 MBY86622.1 GECZ01024299 GECZ01010871 GECZ01008259 GECZ01004068 JAS45470.1 JAS58898.1 JAS61510.1 JAS65701.1 CVRI01000036 CRK92989.1 ABLF02039643 ABLF02039644 ABLF02057225 GGMR01013207 MBY25826.1 CH479199 EDW29517.1 GGFM01006109 MBW26860.1 ADMH02000523 ETN66115.1 GGFJ01007534 MBW56675.1 GFDL01006428 JAV28617.1 UFQS01000322 UFQT01000322 SSX02869.1 SSX23236.1 GFDL01006299 JAV28746.1 GGFK01002467 MBW35788.1 UFQS01001198 UFQT01001198 SSX09651.1 SSX29447.1 GFDL01006431 JAV28614.1 GAPW01002669 JAC10929.1 GAPW01002670 JAC10928.1 JXUM01068902 KQ562524 KXJ75702.1 AXCN02000935 KZ288189 PBC34658.1 AAAB01008960 EAA11146.3 KK107139 QOIP01000003 EZA57891.1 RLU24375.1 APCN01004377 AXCM01004374 DS232312 EDS39824.1 ATLV01011192 KE524650 KFB35932.1 GAPW01002983 JAC10615.1 DS232080 EDS34209.1 KQ979608 KYN20279.1 EDS39823.1 KQ977643 KYN00870.1 KQ981836 KYN35055.1 KQ976542 KYM81139.1 KQ982482 KYQ55848.1 CAQQ02092546 DS235053 EEB11075.1 KQ434827 KZC07560.1
Proteomes
UP000007151
UP000053268
UP000095301
UP000030742
UP000037069
UP000095300
+ More
UP000007266 UP000091820 UP000000304 UP000192221 UP000000803 UP000001292 UP000001819 UP000002282 UP000007801 UP000008711 UP000008792 UP000092461 UP000268350 UP000001070 UP000092553 UP000007798 UP000092462 UP000078200 UP000092445 UP000183832 UP000007819 UP000008744 UP000000673 UP000069272 UP000069940 UP000249989 UP000092443 UP000075884 UP000075900 UP000002358 UP000075920 UP000075886 UP000075880 UP000242457 UP000007062 UP000053097 UP000279307 UP000075881 UP000075903 UP000075840 UP000075883 UP000002320 UP000030765 UP000078492 UP000078542 UP000005203 UP000078541 UP000078540 UP000075809 UP000075901 UP000015102 UP000009046 UP000076502 UP000075885
UP000007266 UP000091820 UP000000304 UP000192221 UP000000803 UP000001292 UP000001819 UP000002282 UP000007801 UP000008711 UP000008792 UP000092461 UP000268350 UP000001070 UP000092553 UP000007798 UP000092462 UP000078200 UP000092445 UP000183832 UP000007819 UP000008744 UP000000673 UP000069272 UP000069940 UP000249989 UP000092443 UP000075884 UP000075900 UP000002358 UP000075920 UP000075886 UP000075880 UP000242457 UP000007062 UP000053097 UP000279307 UP000075881 UP000075903 UP000075840 UP000075883 UP000002320 UP000030765 UP000078492 UP000078542 UP000005203 UP000078541 UP000078540 UP000075809 UP000075901 UP000015102 UP000009046 UP000076502 UP000075885
PRIDE
Pfam
PF10242 L_HMGIC_fpl
Interpro
IPR019372
LHFPL
ProteinModelPortal
A0A2W1B457
A0A212FK78
A0A0N1IE54
A0A1I8MDT8
A0A1Y1LP15
U4URT7
+ More
A0A0L0CKB6 A0A1I8NWS6 D2A1J6 A0A1A9X0Z5 B4QPE3 A0A1W4VS70 Q9VVN1 B4IFP8 Q2LZ07 B4PJF2 A0A0P8ZTY6 B3ND98 B4LF71 B3M3K4 A0A1B0CAN6 W8C5D0 A0A3B0J5F8 U5EPU8 B4J3A4 A0A0M3QX50 B4MLG6 A0A1B0DJ96 A0A1L8D9Y5 A0A0K8UBW9 A0A034V6W1 A0A069DSM9 A0A146KVN9 A0A1B6DRN9 A0A0A9W3R8 A0A1A9UM44 A0A1A9ZDL1 A0A1B6HHV3 A0A2S2RB87 A0A1B6G8W7 A0A1J1HY90 X1WJ41 A0A2S2PA68 B4GZU0 A0A2M3ZEK6 W5JT44 A0A2M4BUD3 A0A1Q3FM42 A0A336LYS8 A0A1Q3FMN7 A0A2M4A566 A0A336KY36 A0A1Q3FM64 A0A182FHN0 A0A023EQP5 A0A023END2 A0A182GJY6 A0A1A9Y1N4 A0A182NMB8 A0A182RC11 K7ISB0 A0A182WLV2 A0A182QDN6 A0A182JJ00 A0A2A3ESF2 Q7Q790 A0A026WQC0 A0A182K069 A0A182URG7 A0A182HM76 A0A182MUD6 B0X3D2 A0A084VD88 A0A023ENU8 B0WT66 A0A195E5Z9 B0X3D1 A0A151IH69 A0A088AGX2 A0A195F3A1 A0A195BAC1 A0A151X6E0 A0A182T0Y4 T1GF29 E0VCG9 A0A154P8U3 A0A182PVV0
A0A0L0CKB6 A0A1I8NWS6 D2A1J6 A0A1A9X0Z5 B4QPE3 A0A1W4VS70 Q9VVN1 B4IFP8 Q2LZ07 B4PJF2 A0A0P8ZTY6 B3ND98 B4LF71 B3M3K4 A0A1B0CAN6 W8C5D0 A0A3B0J5F8 U5EPU8 B4J3A4 A0A0M3QX50 B4MLG6 A0A1B0DJ96 A0A1L8D9Y5 A0A0K8UBW9 A0A034V6W1 A0A069DSM9 A0A146KVN9 A0A1B6DRN9 A0A0A9W3R8 A0A1A9UM44 A0A1A9ZDL1 A0A1B6HHV3 A0A2S2RB87 A0A1B6G8W7 A0A1J1HY90 X1WJ41 A0A2S2PA68 B4GZU0 A0A2M3ZEK6 W5JT44 A0A2M4BUD3 A0A1Q3FM42 A0A336LYS8 A0A1Q3FMN7 A0A2M4A566 A0A336KY36 A0A1Q3FM64 A0A182FHN0 A0A023EQP5 A0A023END2 A0A182GJY6 A0A1A9Y1N4 A0A182NMB8 A0A182RC11 K7ISB0 A0A182WLV2 A0A182QDN6 A0A182JJ00 A0A2A3ESF2 Q7Q790 A0A026WQC0 A0A182K069 A0A182URG7 A0A182HM76 A0A182MUD6 B0X3D2 A0A084VD88 A0A023ENU8 B0WT66 A0A195E5Z9 B0X3D1 A0A151IH69 A0A088AGX2 A0A195F3A1 A0A195BAC1 A0A151X6E0 A0A182T0Y4 T1GF29 E0VCG9 A0A154P8U3 A0A182PVV0
Ontologies
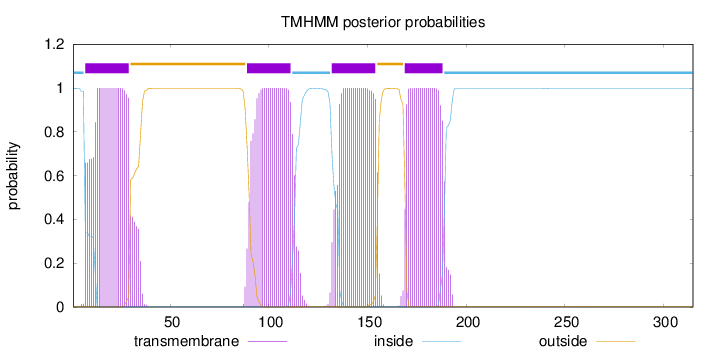
Topology
Length:
315
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
88.6053900000001
Exp number, first 60 AAs:
23.37183
Total prob of N-in:
0.99856
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 88
TMhelix
89 - 111
inside
112 - 131
TMhelix
132 - 154
outside
155 - 168
TMhelix
169 - 188
inside
189 - 315
Population Genetic Test Statistics
Pi
244.329347
Theta
190.463688
Tajima's D
0.966284
CLR
0.337403
CSRT
0.651667416629168
Interpretation
Uncertain
