Gene
KWMTBOMO08376
Pre Gene Modal
BGIBMGA009286
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_Dbp45A_[Amyelois_transitella]
Full name
Probable ATP-dependent RNA helicase Dbp45A
Location in the cell
Nuclear Reliability : 2.604
Sequence
CDS
ATGACCGAGAATGATGGAAAGGAATTTGCTGTGTTAGGAGTAAAACCATGGCTTATTAAACAATTGCTTACGTTGGGTATTAGAACACCAACTCCAATCCAAAAAGGCTGTATATCGCGATTATTGGCTGGTGATGACTGCATTGGAGCTGCCAAAACTGGGTCAGGAAAGACATTTGCATTTGCCCTACCCATTATCCAGCACTTGGCCGAGGATCCATATGGGATTTTTGCATTGGTTCTTACTCCAACACATGAGCTGGCTTATCAGATTGCAGATCAATTCACAATTTTAGGACAACCCCTAAAACTCAGAGTTTGCATAGTGACAGGAGGATCGGATCAAATAGAAGAGTCACTGAAATTAGCTAAAAGACCACATATAGTGGTAGCGATGCCAGGTCGTTTAGCAGATCACATATCAGGTTGTGATACATTCTCATTAAAGAAAATTCAATATTTGGTTCTGGATGAGGCTGATAGGTTATTCAGTGAATCTTTTCAAGAAGATTTAGAAACAATATTCAGTGCTTTACCATCTAAAAGGCAGAATCTTCTTTTCTCAGCTACCATTACACCTGATGTGCGTGAATCGAAATTATTACCCTTAAATAATGATAAACTCCTTGTATGGAAAGAAACTGATGAACAACTAACAGTTTCGACATTGGACCAGCGCTACGTCGTTTGTCCTGCGTACGCCCGTGATGTGTATCTTGTGCAAACTCTGAGGAAATATCGAGAAACAAAACCAAGCTCTCACATCATTGTCTTTACTGATACAAAGAAAGAATGTCAAGTCCTTTCGATGATGTTAAACGCGATATCGATGGACAATATTTGTCTTCACGGATTTATGCGCCAGAAGGAACGCGTGGCAGCTTTAGCGCAGTTCCGGAGCAACCTCAAATGCACCTTGGTGGCCACGAACGTGGCCGCTAGGGGCTTGGACATACCAACCGTGGACCTCGTCGTCAACCACAAACTACCGCTCGAGCCTAAGGAATATATACATAGGGTCGGCAGAACCGCCAGAGCAGGTAGAAGCGGAATGGCGATATCTCTGATAACACCCTACGACATTCTGCGCCTCGGAGAAATCGAAGATCAGATCAAGACTAAACTGACTGAATACAAAATCGACGACGACGAAGCTGTGAAGGTGTTCACGACGGTGAGCGTGACGCGCCGCGAACAAGAGGCGCAGCTTGACAACGAACAGTTTGAACAGCGGAAACGCAGCTACAGGGTCAAGAGGTGGATCCAAGCTGGCGTCAATCCGTTGTTTATGGAGCAGGGGTTAGAAGAAGCGCGGAGACAACGTATTAAAGCCGCGAAGAAAGCGAAACTCGCCAAAATTAAGAATATACAGTCACAGATGAACGAAGACAAATCAGATGTGAACACCGAGGAACGCCTGCGACCCTGCGAAGACAAGGACATCGATAGCACTATCAAAGAAGGCCTAAAGAAAGTCAACAAAAGGCTAATGAAACAAGACGAACGCTTTAAAAATGTTATAGGAAAGATTACTAAAATAAAACACAGAGCCAACAAATTGCAGGCCAAGCAGGAAGCAGAAGACATGGCTCCGAAGAAAAAGAAAAAGAAAAATGTCAAATAA
Protein
MTENDGKEFAVLGVKPWLIKQLLTLGIRTPTPIQKGCISRLLAGDDCIGAAKTGSGKTFAFALPIIQHLAEDPYGIFALVLTPTHELAYQIADQFTILGQPLKLRVCIVTGGSDQIEESLKLAKRPHIVVAMPGRLADHISGCDTFSLKKIQYLVLDEADRLFSESFQEDLETIFSALPSKRQNLLFSATITPDVRESKLLPLNNDKLLVWKETDEQLTVSTLDQRYVVCPAYARDVYLVQTLRKYRETKPSSHIIVFTDTKKECQVLSMMLNAISMDNICLHGFMRQKERVAALAQFRSNLKCTLVATNVAARGLDIPTVDLVVNHKLPLEPKEYIHRVGRTARAGRSGMAISLITPYDILRLGEIEDQIKTKLTEYKIDDDEAVKVFTTVSVTRREQEAQLDNEQFEQRKRSYRVKRWIQAGVNPLFMEQGLEEARRQRIKAAKKAKLAKIKNIQSQMNEDKSDVNTEERLRPCEDKDIDSTIKEGLKKVNKRLMKQDERFKNVIGKITKIKHRANKLQAKQEAEDMAPKKKKKKNVK
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the DEAD box helicase family.
Belongs to the DEAD box helicase family. DDX49/DBP8 subfamily.
Belongs to the DEAD box helicase family. DDX49/DBP8 subfamily.
Keywords
ATP-binding
Complete proteome
Helicase
Hydrolase
Nucleotide-binding
Reference proteome
RNA-binding
Feature
chain Probable ATP-dependent RNA helicase Dbp45A
Uniprot
H9JID6
A0A2H1VJ25
A0A2W1BKZ0
S4P814
A0A437BQ63
A0A212F3G5
+ More
A0A194PYZ9 A0A0N0PAV1 W8B7M3 Q16F43 A0A3B0JTN4 A0A034WSI6 A0A1Q3EWQ8 A0A0A1WLG5 A0A0K8WBQ5 B0WPH6 A0A0L0C6Z2 B3N814 A0A1S4G323 A0A1I8Q7K1 A0A1B0CK84 B4GHY6 B4P3W4 A0A1I8M2B7 Q28ZC3 B4NN30 Q07886 B4HSL4 B4QGR3 A0A1W4UL26 B3MI80 A0A1B0BYF6 B4LM05 B4J5B0 B4KLK7 A0A182H8H6 A0A1A9WNI9 A0A084WLA7 A0A182N802 W5JRI5 A0A336MCY9 A0A2M4ARC0 A0A182YNR9 A0A2M4ARA1 A0A0M4E4S9 A0A182FI17 A0A182PHE3 A0A182RTP9 A0A2M4BKB9 A0A2M4BK69 A0A2M3Z790 A0A182WAB5 A0A182K139 A0A182UTC9 A0A182HP60 Q7Q9N8 A0A182LBD3 A0A182XJA2 A0A182TX35 A0A0C9R257 A0A154PL82 E2AZ53 A0A0L7QL12 D2A1T9 A0A0J7LA51 A0A067QV09 A0A2J7PG42 A0A088AUP0 A0A3L8DV89 K7IZH6 A0A232ESA4 A0A2M3Z848 A0A2A3ER93 A0A2P8Y084 E9IKL6 A0A195DR50 A0A0P5Y4U3 A0A0P5SD39 A0A0P5XJ24 A0A0P5ZB71 A0A1B6EXN9 E9GSH7 A0A151K2N1 A0A0N8DP27 A0A0P5NYN8 A0A1B6L5D5 A0A0P5JQ86 A0A0P5WNR9 F4X8M8 A0A0M8ZU72 A0A158NBJ6 A0A195FCW8 A0A1J1HXT4 A0A1Y1N604 A0A3Q0JIL3 A0A1B6CLD4 E2C3Z0 A0A0P5LTD3 A0A026W4Q2 A0A1L8DG04
A0A194PYZ9 A0A0N0PAV1 W8B7M3 Q16F43 A0A3B0JTN4 A0A034WSI6 A0A1Q3EWQ8 A0A0A1WLG5 A0A0K8WBQ5 B0WPH6 A0A0L0C6Z2 B3N814 A0A1S4G323 A0A1I8Q7K1 A0A1B0CK84 B4GHY6 B4P3W4 A0A1I8M2B7 Q28ZC3 B4NN30 Q07886 B4HSL4 B4QGR3 A0A1W4UL26 B3MI80 A0A1B0BYF6 B4LM05 B4J5B0 B4KLK7 A0A182H8H6 A0A1A9WNI9 A0A084WLA7 A0A182N802 W5JRI5 A0A336MCY9 A0A2M4ARC0 A0A182YNR9 A0A2M4ARA1 A0A0M4E4S9 A0A182FI17 A0A182PHE3 A0A182RTP9 A0A2M4BKB9 A0A2M4BK69 A0A2M3Z790 A0A182WAB5 A0A182K139 A0A182UTC9 A0A182HP60 Q7Q9N8 A0A182LBD3 A0A182XJA2 A0A182TX35 A0A0C9R257 A0A154PL82 E2AZ53 A0A0L7QL12 D2A1T9 A0A0J7LA51 A0A067QV09 A0A2J7PG42 A0A088AUP0 A0A3L8DV89 K7IZH6 A0A232ESA4 A0A2M3Z848 A0A2A3ER93 A0A2P8Y084 E9IKL6 A0A195DR50 A0A0P5Y4U3 A0A0P5SD39 A0A0P5XJ24 A0A0P5ZB71 A0A1B6EXN9 E9GSH7 A0A151K2N1 A0A0N8DP27 A0A0P5NYN8 A0A1B6L5D5 A0A0P5JQ86 A0A0P5WNR9 F4X8M8 A0A0M8ZU72 A0A158NBJ6 A0A195FCW8 A0A1J1HXT4 A0A1Y1N604 A0A3Q0JIL3 A0A1B6CLD4 E2C3Z0 A0A0P5LTD3 A0A026W4Q2 A0A1L8DG04
EC Number
3.6.4.13
Pubmed
19121390
28756777
23622113
22118469
26354079
24495485
+ More
17510324 25348373 25830018 26108605 17994087 17550304 25315136 15632085 7692973 10731132 12537572 12537569 22936249 26483478 24438588 20920257 23761445 25244985 12364791 14747013 17210077 20966253 20798317 18362917 19820115 24845553 30249741 20075255 28648823 29403074 21282665 21292972 21719571 21347285 28004739 24508170
17510324 25348373 25830018 26108605 17994087 17550304 25315136 15632085 7692973 10731132 12537572 12537569 22936249 26483478 24438588 20920257 23761445 25244985 12364791 14747013 17210077 20966253 20798317 18362917 19820115 24845553 30249741 20075255 28648823 29403074 21282665 21292972 21719571 21347285 28004739 24508170
EMBL
BABH01022350
BABH01022351
ODYU01002829
SOQ40796.1
KZ150057
PZC74304.1
+ More
GAIX01004349 JAA88211.1 RSAL01000021 RVE52543.1 AGBW02010576 OWR48253.1 KQ459585 KPI98263.1 KQ461193 KPJ07012.1 GAMC01011968 JAB94587.1 CH478514 EAT32857.1 OUUW01000001 SPP74438.1 GAKP01000406 JAC58546.1 GFDL01015296 JAV19749.1 GBXI01014957 JAC99334.1 GDHF01003686 JAI48628.1 DS232024 EDS32364.1 JRES01000930 KNC27204.1 CH954177 EDV59427.1 AJWK01015894 CH479183 EDW36106.1 CM000157 EDW89447.1 CM000071 EAL25690.3 CH964282 EDW85769.1 L13612 Z23266 AE013599 AY058728 CH480816 EDW47041.1 CM000362 CM002911 EDX06288.1 KMY92422.1 CH902619 EDV35925.1 JXJN01022628 JXJN01022629 JXJN01022630 CH940648 EDW59925.1 CH916367 EDW01752.1 CH933808 EDW08643.1 JXUM01118492 KQ566193 KXJ70333.1 ATLV01024210 KE525350 KFB51001.1 ADMH02000632 ETN65525.1 UFQT01000392 SSX23898.1 GGFK01010002 MBW43323.1 GGFK01009982 MBW43303.1 CP012524 ALC41141.1 GGFJ01004282 MBW53423.1 GGFJ01004281 MBW53422.1 GGFM01003645 MBW24396.1 APCN01002921 AAAB01008900 EAA09455.3 GBYB01002085 JAG71852.1 KQ434954 KZC12616.1 GL444081 EFN61290.1 KQ414934 KOC59259.1 KQ971338 EFA02088.1 LBMM01000150 KMR04723.1 KK852918 KDR13848.1 NEVH01025635 PNF15302.1 QOIP01000004 RLU23829.1 NNAY01002453 OXU21243.1 GGFM01003943 MBW24694.1 KZ288192 PBC34227.1 PYGN01001092 PSN37672.1 GL764015 EFZ18887.1 KQ980581 KYN15390.1 GDIP01062952 GDIP01044566 LRGB01000190 JAM40763.1 KZS20479.1 GDIP01141606 JAL62108.1 GDIP01083648 JAM20067.1 GDIP01046112 JAM57603.1 GECZ01026973 JAS42796.1 GL732562 EFX77426.1 LKEX01017577 KYN50273.1 GDIP01014831 JAM88884.1 GDIQ01135983 JAL15743.1 GEBQ01021136 JAT18841.1 GDIQ01195502 JAK56223.1 GDIP01083649 JAM20066.1 GL888932 EGI57297.1 KQ435883 KOX69693.1 ADTU01011121 KQ981673 KYN38253.1 CVRI01000035 CRK92896.1 GEZM01013806 JAV92320.1 GEDC01023066 JAS14232.1 GL452364 EFN77376.1 GDIQ01165431 JAK86294.1 KK107453 EZA50571.1 GFDF01008723 JAV05361.1
GAIX01004349 JAA88211.1 RSAL01000021 RVE52543.1 AGBW02010576 OWR48253.1 KQ459585 KPI98263.1 KQ461193 KPJ07012.1 GAMC01011968 JAB94587.1 CH478514 EAT32857.1 OUUW01000001 SPP74438.1 GAKP01000406 JAC58546.1 GFDL01015296 JAV19749.1 GBXI01014957 JAC99334.1 GDHF01003686 JAI48628.1 DS232024 EDS32364.1 JRES01000930 KNC27204.1 CH954177 EDV59427.1 AJWK01015894 CH479183 EDW36106.1 CM000157 EDW89447.1 CM000071 EAL25690.3 CH964282 EDW85769.1 L13612 Z23266 AE013599 AY058728 CH480816 EDW47041.1 CM000362 CM002911 EDX06288.1 KMY92422.1 CH902619 EDV35925.1 JXJN01022628 JXJN01022629 JXJN01022630 CH940648 EDW59925.1 CH916367 EDW01752.1 CH933808 EDW08643.1 JXUM01118492 KQ566193 KXJ70333.1 ATLV01024210 KE525350 KFB51001.1 ADMH02000632 ETN65525.1 UFQT01000392 SSX23898.1 GGFK01010002 MBW43323.1 GGFK01009982 MBW43303.1 CP012524 ALC41141.1 GGFJ01004282 MBW53423.1 GGFJ01004281 MBW53422.1 GGFM01003645 MBW24396.1 APCN01002921 AAAB01008900 EAA09455.3 GBYB01002085 JAG71852.1 KQ434954 KZC12616.1 GL444081 EFN61290.1 KQ414934 KOC59259.1 KQ971338 EFA02088.1 LBMM01000150 KMR04723.1 KK852918 KDR13848.1 NEVH01025635 PNF15302.1 QOIP01000004 RLU23829.1 NNAY01002453 OXU21243.1 GGFM01003943 MBW24694.1 KZ288192 PBC34227.1 PYGN01001092 PSN37672.1 GL764015 EFZ18887.1 KQ980581 KYN15390.1 GDIP01062952 GDIP01044566 LRGB01000190 JAM40763.1 KZS20479.1 GDIP01141606 JAL62108.1 GDIP01083648 JAM20067.1 GDIP01046112 JAM57603.1 GECZ01026973 JAS42796.1 GL732562 EFX77426.1 LKEX01017577 KYN50273.1 GDIP01014831 JAM88884.1 GDIQ01135983 JAL15743.1 GEBQ01021136 JAT18841.1 GDIQ01195502 JAK56223.1 GDIP01083649 JAM20066.1 GL888932 EGI57297.1 KQ435883 KOX69693.1 ADTU01011121 KQ981673 KYN38253.1 CVRI01000035 CRK92896.1 GEZM01013806 JAV92320.1 GEDC01023066 JAS14232.1 GL452364 EFN77376.1 GDIQ01165431 JAK86294.1 KK107453 EZA50571.1 GFDF01008723 JAV05361.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000008820
+ More
UP000268350 UP000002320 UP000037069 UP000008711 UP000095300 UP000092461 UP000008744 UP000002282 UP000095301 UP000001819 UP000007798 UP000000803 UP000001292 UP000000304 UP000192221 UP000007801 UP000092460 UP000008792 UP000001070 UP000009192 UP000069940 UP000249989 UP000091820 UP000030765 UP000075884 UP000000673 UP000076408 UP000092553 UP000069272 UP000075885 UP000075900 UP000075920 UP000075881 UP000075903 UP000075840 UP000007062 UP000075882 UP000076407 UP000075902 UP000076502 UP000000311 UP000053825 UP000007266 UP000036403 UP000027135 UP000235965 UP000005203 UP000279307 UP000002358 UP000215335 UP000242457 UP000245037 UP000078492 UP000076858 UP000000305 UP000078542 UP000007755 UP000053105 UP000005205 UP000078541 UP000183832 UP000079169 UP000008237 UP000053097
UP000268350 UP000002320 UP000037069 UP000008711 UP000095300 UP000092461 UP000008744 UP000002282 UP000095301 UP000001819 UP000007798 UP000000803 UP000001292 UP000000304 UP000192221 UP000007801 UP000092460 UP000008792 UP000001070 UP000009192 UP000069940 UP000249989 UP000091820 UP000030765 UP000075884 UP000000673 UP000076408 UP000092553 UP000069272 UP000075885 UP000075900 UP000075920 UP000075881 UP000075903 UP000075840 UP000007062 UP000075882 UP000076407 UP000075902 UP000076502 UP000000311 UP000053825 UP000007266 UP000036403 UP000027135 UP000235965 UP000005203 UP000279307 UP000002358 UP000215335 UP000242457 UP000245037 UP000078492 UP000076858 UP000000305 UP000078542 UP000007755 UP000053105 UP000005205 UP000078541 UP000183832 UP000079169 UP000008237 UP000053097
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9JID6
A0A2H1VJ25
A0A2W1BKZ0
S4P814
A0A437BQ63
A0A212F3G5
+ More
A0A194PYZ9 A0A0N0PAV1 W8B7M3 Q16F43 A0A3B0JTN4 A0A034WSI6 A0A1Q3EWQ8 A0A0A1WLG5 A0A0K8WBQ5 B0WPH6 A0A0L0C6Z2 B3N814 A0A1S4G323 A0A1I8Q7K1 A0A1B0CK84 B4GHY6 B4P3W4 A0A1I8M2B7 Q28ZC3 B4NN30 Q07886 B4HSL4 B4QGR3 A0A1W4UL26 B3MI80 A0A1B0BYF6 B4LM05 B4J5B0 B4KLK7 A0A182H8H6 A0A1A9WNI9 A0A084WLA7 A0A182N802 W5JRI5 A0A336MCY9 A0A2M4ARC0 A0A182YNR9 A0A2M4ARA1 A0A0M4E4S9 A0A182FI17 A0A182PHE3 A0A182RTP9 A0A2M4BKB9 A0A2M4BK69 A0A2M3Z790 A0A182WAB5 A0A182K139 A0A182UTC9 A0A182HP60 Q7Q9N8 A0A182LBD3 A0A182XJA2 A0A182TX35 A0A0C9R257 A0A154PL82 E2AZ53 A0A0L7QL12 D2A1T9 A0A0J7LA51 A0A067QV09 A0A2J7PG42 A0A088AUP0 A0A3L8DV89 K7IZH6 A0A232ESA4 A0A2M3Z848 A0A2A3ER93 A0A2P8Y084 E9IKL6 A0A195DR50 A0A0P5Y4U3 A0A0P5SD39 A0A0P5XJ24 A0A0P5ZB71 A0A1B6EXN9 E9GSH7 A0A151K2N1 A0A0N8DP27 A0A0P5NYN8 A0A1B6L5D5 A0A0P5JQ86 A0A0P5WNR9 F4X8M8 A0A0M8ZU72 A0A158NBJ6 A0A195FCW8 A0A1J1HXT4 A0A1Y1N604 A0A3Q0JIL3 A0A1B6CLD4 E2C3Z0 A0A0P5LTD3 A0A026W4Q2 A0A1L8DG04
A0A194PYZ9 A0A0N0PAV1 W8B7M3 Q16F43 A0A3B0JTN4 A0A034WSI6 A0A1Q3EWQ8 A0A0A1WLG5 A0A0K8WBQ5 B0WPH6 A0A0L0C6Z2 B3N814 A0A1S4G323 A0A1I8Q7K1 A0A1B0CK84 B4GHY6 B4P3W4 A0A1I8M2B7 Q28ZC3 B4NN30 Q07886 B4HSL4 B4QGR3 A0A1W4UL26 B3MI80 A0A1B0BYF6 B4LM05 B4J5B0 B4KLK7 A0A182H8H6 A0A1A9WNI9 A0A084WLA7 A0A182N802 W5JRI5 A0A336MCY9 A0A2M4ARC0 A0A182YNR9 A0A2M4ARA1 A0A0M4E4S9 A0A182FI17 A0A182PHE3 A0A182RTP9 A0A2M4BKB9 A0A2M4BK69 A0A2M3Z790 A0A182WAB5 A0A182K139 A0A182UTC9 A0A182HP60 Q7Q9N8 A0A182LBD3 A0A182XJA2 A0A182TX35 A0A0C9R257 A0A154PL82 E2AZ53 A0A0L7QL12 D2A1T9 A0A0J7LA51 A0A067QV09 A0A2J7PG42 A0A088AUP0 A0A3L8DV89 K7IZH6 A0A232ESA4 A0A2M3Z848 A0A2A3ER93 A0A2P8Y084 E9IKL6 A0A195DR50 A0A0P5Y4U3 A0A0P5SD39 A0A0P5XJ24 A0A0P5ZB71 A0A1B6EXN9 E9GSH7 A0A151K2N1 A0A0N8DP27 A0A0P5NYN8 A0A1B6L5D5 A0A0P5JQ86 A0A0P5WNR9 F4X8M8 A0A0M8ZU72 A0A158NBJ6 A0A195FCW8 A0A1J1HXT4 A0A1Y1N604 A0A3Q0JIL3 A0A1B6CLD4 E2C3Z0 A0A0P5LTD3 A0A026W4Q2 A0A1L8DG04
PDB
5IVL
E-value=9.60332e-57,
Score=559
Ontologies
GO
Topology
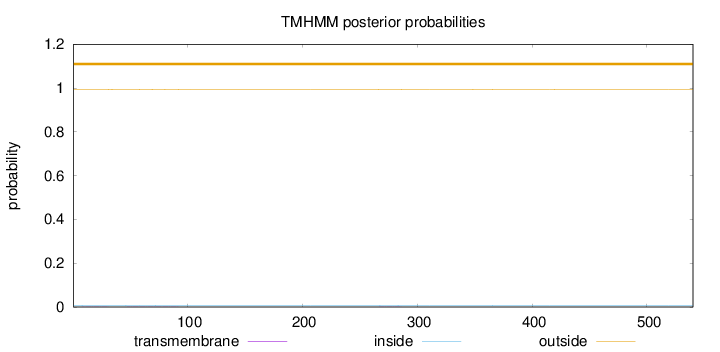
Length:
540
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0475600000000001
Exp number, first 60 AAs:
0.01721
Total prob of N-in:
0.00782
outside
1 - 540
Population Genetic Test Statistics
Pi
244.795666
Theta
199.04226
Tajima's D
0.716934
CLR
0.675447
CSRT
0.578971051447428
Interpretation
Uncertain
