Gene
KWMTBOMO08375
Pre Gene Modal
BGIBMGA009218
Annotation
PREDICTED:_uncharacterized_protein_LOC106105586_[Papilio_polytes]
Location in the cell
Extracellular Reliability : 1.058 Mitochondrial Reliability : 1.06
Sequence
CDS
ATGAAACTGGACCAGCAAAGAGACAGGCCTCTCTGCATAATGATCAACTGGCTGCTGGCCCGACAGAAGCACGTTATGAAATACGCGACGTTGTACTTGGAGCAGGGGTTCGACGTGCTCTCGGTGTCGTGCACTCCGTGGCAGCTAATGTGGCCCATGAAGGGATCACAGCTAGTTGCCGGTGACGTTCTCAAATTCATGGCTTCGAACGAGAATGAGCAGCCCCTCGTGGTGCACGGGTTCTCGATTGCTGGATACATGTGGGGAGAAGTTTACGTGCACGTCATGAATAATAGGAAGATGTACCAACCGGTAATGGACCGCATCGCGGCGCAGGTGTGGGACTCTGCCGCGGACATCAGCGAGATCACGATCGGCGTGCCGGCCGCCGTCTTCCCGAAGAACCAGATCATGCAGAATACTCTGCGGGCGTATATGGAGTGA
Protein
MKLDQQRDRPLCIMINWLLARQKHVMKYATLYLEQGFDVLSVSCTPWQLMWPMKGSQLVAGDVLKFMASNENEQPLVVHGFSIAGYMWGEVYVHVMNNRKMYQPVMDRIAAQVWDSAADISEITIGVPAAVFPKNQIMQNTLRAYME
Summary
Uniprot
H9JI70
A0A2A4JBY1
A0A2H1VIY2
S4P3U4
A0A212F3F7
A0A1E1WGU2
+ More
A0A0N1PHU4 A0A194PY52 A0A2W1BGW5 A0A437BQ88 A0A0T6BCD8 A0A2W1BJ24 A0A2A4JAE8 A0A2A4IXV0 A0A3S2M6I1 H9JI72 A0A336MIF2 A0A336L0G7 A0A084WBD8 V5I9V6 W5JQH1 A0A2M4BTJ8 A0A2M4AA03 A0A182U739 A0A2M3Z4F8 T1E9J9 A0A182FZY1 A0A2M3Z4H2 A0A2M3Z4G1 Q5TPV0 A0A2M3Z4G6 A0A2M4BPV3 A0A194PY42 A0A2M4BNZ2 A0A182IQZ4 D2A377 A0A182Q2W6 A0A182VIU1 A0A182XC95 A0A182HNJ6 A0A1S4H6S2 A0A182MXV8 A0A182T4N6 A0A067QWY7 A0A1Y9IW18 A0A195BAK3 A0A158NBJ4 A0A182LPH8 F4X8M9 A0A151K2H1 A0A195DRC3 A0A026W3B8 A0A1W4X2A0 A0A182R8P5 A0A2J7PG46 T1DD53 A0A195FDH0 A0A088AV16 A0A182JPA9 A0A182P9C0 Q16XW6 A0A0K8TP18 A0A182YPE9 N6UV01 A0A0L7QKZ7 A0A182GTT0 A0A023ET29 A0A1J1I7T0 A0A154PL67 A0A1B6EPV0 A0A1L8E854 A0A1L8E932 A0A1L8EA50 A0A1L8EDM5 T1E249 A0A0K8WHH9 A0A1B6MUY4 A0A034WBB8 A0A1B0CEJ9 A0A1B0EY32 A0A226E7W4 A0A182GTT1 A0A0K8UE87 A0A1L8DC40 B0WGX1 A0A0A1XJ14 A0A1Q3FK97 V9ICC0 A0A1Q3FK98 A0A1Y1KS93 A0A1I8P0T5 A0A232ESC2 A0A1I8P0W5 K7IZH7 A0A0A1XP76 A0A0P8YDC9 A0A0R1DTM0 V9IE41 A0A1A9WHX6 A0A1I8P0T0
A0A0N1PHU4 A0A194PY52 A0A2W1BGW5 A0A437BQ88 A0A0T6BCD8 A0A2W1BJ24 A0A2A4JAE8 A0A2A4IXV0 A0A3S2M6I1 H9JI72 A0A336MIF2 A0A336L0G7 A0A084WBD8 V5I9V6 W5JQH1 A0A2M4BTJ8 A0A2M4AA03 A0A182U739 A0A2M3Z4F8 T1E9J9 A0A182FZY1 A0A2M3Z4H2 A0A2M3Z4G1 Q5TPV0 A0A2M3Z4G6 A0A2M4BPV3 A0A194PY42 A0A2M4BNZ2 A0A182IQZ4 D2A377 A0A182Q2W6 A0A182VIU1 A0A182XC95 A0A182HNJ6 A0A1S4H6S2 A0A182MXV8 A0A182T4N6 A0A067QWY7 A0A1Y9IW18 A0A195BAK3 A0A158NBJ4 A0A182LPH8 F4X8M9 A0A151K2H1 A0A195DRC3 A0A026W3B8 A0A1W4X2A0 A0A182R8P5 A0A2J7PG46 T1DD53 A0A195FDH0 A0A088AV16 A0A182JPA9 A0A182P9C0 Q16XW6 A0A0K8TP18 A0A182YPE9 N6UV01 A0A0L7QKZ7 A0A182GTT0 A0A023ET29 A0A1J1I7T0 A0A154PL67 A0A1B6EPV0 A0A1L8E854 A0A1L8E932 A0A1L8EA50 A0A1L8EDM5 T1E249 A0A0K8WHH9 A0A1B6MUY4 A0A034WBB8 A0A1B0CEJ9 A0A1B0EY32 A0A226E7W4 A0A182GTT1 A0A0K8UE87 A0A1L8DC40 B0WGX1 A0A0A1XJ14 A0A1Q3FK97 V9ICC0 A0A1Q3FK98 A0A1Y1KS93 A0A1I8P0T5 A0A232ESC2 A0A1I8P0W5 K7IZH7 A0A0A1XP76 A0A0P8YDC9 A0A0R1DTM0 V9IE41 A0A1A9WHX6 A0A1I8P0T0
Pubmed
EMBL
BABH01022349
NWSH01002192
PCG68942.1
ODYU01002829
SOQ40795.1
GAIX01011460
+ More
JAA81100.1 AGBW02010576 OWR48252.1 GDQN01004844 JAT86210.1 KQ461193 KPJ07010.1 KQ459585 KPI98262.1 KZ150057 PZC74302.1 RSAL01000021 RVE52544.1 LJIG01002030 KRT84910.1 PZC74301.1 PCG68941.1 NWSH01005065 PCG64469.1 RVE52545.1 BABH01022347 BABH01022348 UFQS01001048 UFQT01001048 SSX08699.1 SSX28613.1 UFQS01001411 UFQT01001411 SSX10745.1 SSX30427.1 ATLV01022344 KE525331 KFB47532.1 GALX01002270 JAB66196.1 ADMH02000409 ETN66662.1 GGFJ01007228 MBW56369.1 GGFK01004251 MBW37572.1 GGFM01002634 MBW23385.1 GAMD01001282 JAB00309.1 GGFM01002668 MBW23419.1 GGFM01002658 MBW23409.1 AAAB01008966 EAL39475.3 GGFM01002653 MBW23404.1 GGFJ01005959 MBW55100.1 KPI98261.1 GGFJ01005659 MBW54800.1 KQ971338 EFA01952.2 AXCN02002115 APCN01001937 KK852918 KDR13849.1 KQ976540 KYM81262.1 ADTU01011121 GL888932 EGI57298.1 LKEX01017577 KYN50272.1 KQ980581 KYN15391.1 KK107453 QOIP01000004 EZA50570.1 RLU23935.1 NEVH01025635 PNF15303.1 GALA01001552 JAA93300.1 KQ981673 KYN38252.1 CH477530 EAT39478.1 GDAI01001514 JAI16089.1 APGK01010787 KB738301 KB632179 ENN82672.1 ERL89606.1 KQ414934 KOC59260.1 JXUM01087786 KQ563658 KXJ73538.1 GAPW01002074 JAC11524.1 CVRI01000043 CRK95794.1 KQ434954 KZC12615.1 GECZ01029804 JAS39965.1 GFDG01004014 JAV14785.1 GFDG01003625 JAV15174.1 GFDG01003356 JAV15443.1 GFDG01002002 JAV16797.1 GALA01001059 JAA93793.1 GDHF01010606 GDHF01001706 JAI41708.1 JAI50608.1 GEBQ01000256 JAT39721.1 GAKP01006091 GAKP01006090 GAKP01006089 JAC52861.1 AJWK01008941 AJWK01008942 AJWK01008943 AJVK01007412 LNIX01000006 OXA53184.1 KXJ73539.1 GDHF01032408 GDHF01027647 GDHF01027436 GDHF01006664 GDHF01004245 JAI19906.1 JAI24667.1 JAI24878.1 JAI45650.1 JAI48069.1 GFDF01010139 JAV03945.1 DS231930 EDS27265.1 GBXI01002973 JAD11319.1 GFDL01007090 JAV27955.1 JR039501 AEY58730.1 GFDL01007111 JAV27934.1 GEZM01075092 JAV64289.1 NNAY01002453 OXU21242.1 GBXI01001602 JAD12690.1 CH902619 KPU76961.1 CM000158 KRK00445.1 JR039502 AEY58731.1
JAA81100.1 AGBW02010576 OWR48252.1 GDQN01004844 JAT86210.1 KQ461193 KPJ07010.1 KQ459585 KPI98262.1 KZ150057 PZC74302.1 RSAL01000021 RVE52544.1 LJIG01002030 KRT84910.1 PZC74301.1 PCG68941.1 NWSH01005065 PCG64469.1 RVE52545.1 BABH01022347 BABH01022348 UFQS01001048 UFQT01001048 SSX08699.1 SSX28613.1 UFQS01001411 UFQT01001411 SSX10745.1 SSX30427.1 ATLV01022344 KE525331 KFB47532.1 GALX01002270 JAB66196.1 ADMH02000409 ETN66662.1 GGFJ01007228 MBW56369.1 GGFK01004251 MBW37572.1 GGFM01002634 MBW23385.1 GAMD01001282 JAB00309.1 GGFM01002668 MBW23419.1 GGFM01002658 MBW23409.1 AAAB01008966 EAL39475.3 GGFM01002653 MBW23404.1 GGFJ01005959 MBW55100.1 KPI98261.1 GGFJ01005659 MBW54800.1 KQ971338 EFA01952.2 AXCN02002115 APCN01001937 KK852918 KDR13849.1 KQ976540 KYM81262.1 ADTU01011121 GL888932 EGI57298.1 LKEX01017577 KYN50272.1 KQ980581 KYN15391.1 KK107453 QOIP01000004 EZA50570.1 RLU23935.1 NEVH01025635 PNF15303.1 GALA01001552 JAA93300.1 KQ981673 KYN38252.1 CH477530 EAT39478.1 GDAI01001514 JAI16089.1 APGK01010787 KB738301 KB632179 ENN82672.1 ERL89606.1 KQ414934 KOC59260.1 JXUM01087786 KQ563658 KXJ73538.1 GAPW01002074 JAC11524.1 CVRI01000043 CRK95794.1 KQ434954 KZC12615.1 GECZ01029804 JAS39965.1 GFDG01004014 JAV14785.1 GFDG01003625 JAV15174.1 GFDG01003356 JAV15443.1 GFDG01002002 JAV16797.1 GALA01001059 JAA93793.1 GDHF01010606 GDHF01001706 JAI41708.1 JAI50608.1 GEBQ01000256 JAT39721.1 GAKP01006091 GAKP01006090 GAKP01006089 JAC52861.1 AJWK01008941 AJWK01008942 AJWK01008943 AJVK01007412 LNIX01000006 OXA53184.1 KXJ73539.1 GDHF01032408 GDHF01027647 GDHF01027436 GDHF01006664 GDHF01004245 JAI19906.1 JAI24667.1 JAI24878.1 JAI45650.1 JAI48069.1 GFDF01010139 JAV03945.1 DS231930 EDS27265.1 GBXI01002973 JAD11319.1 GFDL01007090 JAV27955.1 JR039501 AEY58730.1 GFDL01007111 JAV27934.1 GEZM01075092 JAV64289.1 NNAY01002453 OXU21242.1 GBXI01001602 JAD12690.1 CH902619 KPU76961.1 CM000158 KRK00445.1 JR039502 AEY58731.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000030765 UP000000673 UP000075902 UP000069272 UP000007062 UP000075880 UP000007266 UP000075886 UP000075903 UP000076407 UP000075840 UP000075884 UP000075901 UP000027135 UP000075920 UP000078540 UP000005205 UP000075882 UP000007755 UP000078542 UP000078492 UP000053097 UP000279307 UP000192223 UP000075900 UP000235965 UP000078541 UP000005203 UP000075881 UP000075885 UP000008820 UP000076408 UP000019118 UP000030742 UP000053825 UP000069940 UP000249989 UP000183832 UP000076502 UP000092461 UP000092462 UP000198287 UP000002320 UP000095300 UP000215335 UP000002358 UP000007801 UP000002282 UP000091820
UP000030765 UP000000673 UP000075902 UP000069272 UP000007062 UP000075880 UP000007266 UP000075886 UP000075903 UP000076407 UP000075840 UP000075884 UP000075901 UP000027135 UP000075920 UP000078540 UP000005205 UP000075882 UP000007755 UP000078542 UP000078492 UP000053097 UP000279307 UP000192223 UP000075900 UP000235965 UP000078541 UP000005203 UP000075881 UP000075885 UP000008820 UP000076408 UP000019118 UP000030742 UP000053825 UP000069940 UP000249989 UP000183832 UP000076502 UP000092461 UP000092462 UP000198287 UP000002320 UP000095300 UP000215335 UP000002358 UP000007801 UP000002282 UP000091820
Pfam
PF05705 DUF829
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JI70
A0A2A4JBY1
A0A2H1VIY2
S4P3U4
A0A212F3F7
A0A1E1WGU2
+ More
A0A0N1PHU4 A0A194PY52 A0A2W1BGW5 A0A437BQ88 A0A0T6BCD8 A0A2W1BJ24 A0A2A4JAE8 A0A2A4IXV0 A0A3S2M6I1 H9JI72 A0A336MIF2 A0A336L0G7 A0A084WBD8 V5I9V6 W5JQH1 A0A2M4BTJ8 A0A2M4AA03 A0A182U739 A0A2M3Z4F8 T1E9J9 A0A182FZY1 A0A2M3Z4H2 A0A2M3Z4G1 Q5TPV0 A0A2M3Z4G6 A0A2M4BPV3 A0A194PY42 A0A2M4BNZ2 A0A182IQZ4 D2A377 A0A182Q2W6 A0A182VIU1 A0A182XC95 A0A182HNJ6 A0A1S4H6S2 A0A182MXV8 A0A182T4N6 A0A067QWY7 A0A1Y9IW18 A0A195BAK3 A0A158NBJ4 A0A182LPH8 F4X8M9 A0A151K2H1 A0A195DRC3 A0A026W3B8 A0A1W4X2A0 A0A182R8P5 A0A2J7PG46 T1DD53 A0A195FDH0 A0A088AV16 A0A182JPA9 A0A182P9C0 Q16XW6 A0A0K8TP18 A0A182YPE9 N6UV01 A0A0L7QKZ7 A0A182GTT0 A0A023ET29 A0A1J1I7T0 A0A154PL67 A0A1B6EPV0 A0A1L8E854 A0A1L8E932 A0A1L8EA50 A0A1L8EDM5 T1E249 A0A0K8WHH9 A0A1B6MUY4 A0A034WBB8 A0A1B0CEJ9 A0A1B0EY32 A0A226E7W4 A0A182GTT1 A0A0K8UE87 A0A1L8DC40 B0WGX1 A0A0A1XJ14 A0A1Q3FK97 V9ICC0 A0A1Q3FK98 A0A1Y1KS93 A0A1I8P0T5 A0A232ESC2 A0A1I8P0W5 K7IZH7 A0A0A1XP76 A0A0P8YDC9 A0A0R1DTM0 V9IE41 A0A1A9WHX6 A0A1I8P0T0
A0A0N1PHU4 A0A194PY52 A0A2W1BGW5 A0A437BQ88 A0A0T6BCD8 A0A2W1BJ24 A0A2A4JAE8 A0A2A4IXV0 A0A3S2M6I1 H9JI72 A0A336MIF2 A0A336L0G7 A0A084WBD8 V5I9V6 W5JQH1 A0A2M4BTJ8 A0A2M4AA03 A0A182U739 A0A2M3Z4F8 T1E9J9 A0A182FZY1 A0A2M3Z4H2 A0A2M3Z4G1 Q5TPV0 A0A2M3Z4G6 A0A2M4BPV3 A0A194PY42 A0A2M4BNZ2 A0A182IQZ4 D2A377 A0A182Q2W6 A0A182VIU1 A0A182XC95 A0A182HNJ6 A0A1S4H6S2 A0A182MXV8 A0A182T4N6 A0A067QWY7 A0A1Y9IW18 A0A195BAK3 A0A158NBJ4 A0A182LPH8 F4X8M9 A0A151K2H1 A0A195DRC3 A0A026W3B8 A0A1W4X2A0 A0A182R8P5 A0A2J7PG46 T1DD53 A0A195FDH0 A0A088AV16 A0A182JPA9 A0A182P9C0 Q16XW6 A0A0K8TP18 A0A182YPE9 N6UV01 A0A0L7QKZ7 A0A182GTT0 A0A023ET29 A0A1J1I7T0 A0A154PL67 A0A1B6EPV0 A0A1L8E854 A0A1L8E932 A0A1L8EA50 A0A1L8EDM5 T1E249 A0A0K8WHH9 A0A1B6MUY4 A0A034WBB8 A0A1B0CEJ9 A0A1B0EY32 A0A226E7W4 A0A182GTT1 A0A0K8UE87 A0A1L8DC40 B0WGX1 A0A0A1XJ14 A0A1Q3FK97 V9ICC0 A0A1Q3FK98 A0A1Y1KS93 A0A1I8P0T5 A0A232ESC2 A0A1I8P0W5 K7IZH7 A0A0A1XP76 A0A0P8YDC9 A0A0R1DTM0 V9IE41 A0A1A9WHX6 A0A1I8P0T0
Ontologies
GO
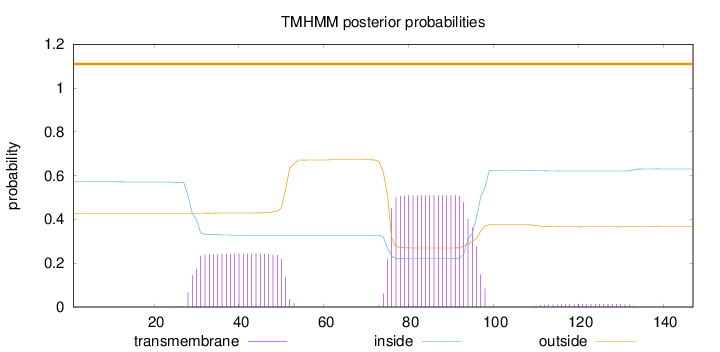
Topology
Length:
147
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
16.349
Exp number, first 60 AAs:
5.41727
Total prob of N-in:
0.57176
outside
1 - 147
Population Genetic Test Statistics
Pi
189.382131
Theta
159.843991
Tajima's D
0.556683
CLR
0.369541
CSRT
0.535623218839058
Interpretation
Uncertain
