Pre Gene Modal
BGIBMGA009222
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_E3_ubiquitin-protein_ligase_UBR1-like_[Bombyx_mori]
Full name
E3 ubiquitin-protein ligase UBR1
Alternative Name
N-recognin
RING-type E3 ubiquitin transferase UBR1
Ubiquitin-protein ligase E3-alpha
RING-type E3 ubiquitin transferase UBR1
Ubiquitin-protein ligase E3-alpha
Location in the cell
Nuclear Reliability : 2.62
Sequence
CDS
ATGAGTACTCCACCGCCAGTGCAGCTGGAGGTCGAGGTTGGTGATGGCTCAATGGATGACGAGACCCCCGACATGATACCCGCCGACAGGTACACGTTGCCGGCTCGCGCTGATGCTGTAGTGGAACTATGGCGGAAGAATCTGCAGGAGGGTGTACTGGCGCCCTCGCACTTTCAAGATCATTGGCGCGTCACCGTTCCGAGAATATATTCGCCACAACCCAACAGAACATGTTTGGACTGGTCTTTCGACGAGGAACTAGCTGCCAAATTGCTCATCCAACCCCTAGAACAGTTCATATGGGGTACAACAGAAGGATCTCAACCGGGTCCCCCTCGTCGCTCTACACTCTGCGGACGGGTGTTCAAACAAGGCGAACCTGCATATAGTTGCAGAGAATGTGGCATGGATAACACTTGCGTACTCTGCGTGGAGTGCTTCAAGGTATCAGCTCATAGACACCACAAGTACAAGATGGGGCAGTCCGGTGGCGGCGGCTGCTGTGACTGCGGTGACGCCGAGGCCTGGAAACGGGATCCATTCTGTGATTTGCATGCTGCTCCAAGCGATCAACCGCCCGAGGCAGGCATAAGCGAGGATGTTATAAGAAGAATGAAGATCGTAGCATCAGCTTGTTTGACTTATTGTCTAAGGCTGCTTACACACGATCACGCTCCATGCTTGCCGAACGATCTCAGATTGAAAGACACGGAGCGCGACATCCTGCAGATACTGGACCAGCCGGATTGTTACTGCACGGTGCTCTACAACGACGAAACTCACACCTTCGAGCAGGTGATAACAACGTTGATACGCGTGACGAAGTGCAACCACCGCGACTCCGTGGAGCTGGTGAGCCTCATCGACCGCGAGGGTCGGGCGCTCATCAAGTGCAACTCGTTCCAGGTCTGCGACAAACTCAAAACGGACATCGAGATGTTCACGGCGCGCCAGAGCTCCACGCTGAAGGTGCTGGTCATGCACGCGCACGTCATCGCGCACCAGACCTTCGCCATGAAGCTGCTCACCTGGCTTCAGAACTTCGTAAGCCAAGAGCAGAGTCTGCGCATGGCCGTGTCCCACGTGGCGCTGGGCGAGGACATGTGGGGCTCGAGCACGGCGGTGGGGGGCGGCGGGGTGGCCGTCGGCGTCATGCAGAACGACTTCCGCATGTGGAAGGCCGCCAGGACCGCCTGGCATCGGCTGCTCATCGCCACCACGCTCATGGACTATTCTACGAAGAGAACCATGGCTATATTATTCACCAAAAATTATGGCTCAATACTGAAGGACTTCATACGAGACGACCACGACCACTCGTTCTCGATATCGTCGCTGTCGGTGCAGCTGTACACGACGCCGACGCTGGCGCACCTGCTCATCGCGAAGCACGACGCGCTCTTCGTCGTGCTCAACACTTTTGTGAGCGAGTGTAACCGACGCTGCAATAATGTGAGCGACACAAGTCATTTCTTAATAAATCTGTCATCGGTGCCCTACGCGGCGCACTGA
Protein
MSTPPPVQLEVEVGDGSMDDETPDMIPADRYTLPARADAVVELWRKNLQEGVLAPSHFQDHWRVTVPRIYSPQPNRTCLDWSFDEELAAKLLIQPLEQFIWGTTEGSQPGPPRRSTLCGRVFKQGEPAYSCRECGMDNTCVLCVECFKVSAHRHHKYKMGQSGGGGCCDCGDAEAWKRDPFCDLHAAPSDQPPEAGISEDVIRRMKIVASACLTYCLRLLTHDHAPCLPNDLRLKDTERDILQILDQPDCYCTVLYNDETHTFEQVITTLIRVTKCNHRDSVELVSLIDREGRALIKCNSFQVCDKLKTDIEMFTARQSSTLKVLVMHAHVIAHQTFAMKLLTWLQNFVSQEQSLRMAVSHVALGEDMWGSSTAVGGGGVAVGVMQNDFRMWKAARTAWHRLLIATTLMDYSTKRTMAILFTKNYGSILKDFIRDDHDHSFSISSLSVQLYTTPTLAHLLIAKHDALFVVLNTFVSECNRRCNNVSDTSHFLINLSSVPYAAH
Summary
Description
E3 ubiquitin-protein ligase which is a component of the N-end rule pathway. Recognizes and binds to proteins bearing specific N-terminal residues that are destabilizing according to the N-end rule, leading to their ubiquitination and subsequent degradation.
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Similarity
Belongs to the UBR1 family.
Keywords
Complete proteome
Metal-binding
Reference proteome
Transferase
Ubl conjugation pathway
Zinc
Zinc-finger
Feature
chain E3 ubiquitin-protein ligase UBR1
Uniprot
A0A3S2LFD2
A0A212F3D3
A0A2W1BLZ8
A0A0N1PGS7
A0A2H1VG67
A0A0C9QG31
+ More
A0A0C9PMM6 A0A2P8Y099 A0A1B6HG01 A0A067QUM0 A0A2J7PG03 A0A232F9W8 K7ISE8 A0A2J7PFZ9 A0A088AH87 A0A2A3ESZ3 E2A1G8 A0A0J7L4E5 E2C439 A0A0M8ZYU1 A0A154P6P7 A0A151K0T7 F4WHK3 A0A158N904 A0A151J379 E9JAU8 A0A1Y1LPA2 A0A151WNN1 A0A1B6DEC1 A0A3L8DSS3 A0A026VTD2 A0A151IBI5 E0VHX5 A0A0L7QWM1 A0A2R7W393 D1ZZM2 A0A146LN83 A0A0A9XCN8 A0A0K8SXL6 A0A1B6EHY3 A0A2H8TIX0 A0A2S2QT54 A0A1B6F6Z8 J9JLI5 N6TQ88 N6ULN5 T1IFX1 N6TMT9 U4UDA6 A0A0P4WDX2 A0A0P4WHX4 A0A087TYA0 A0A224YM26 A0A444SJ90 L7MG75 A0A131YJ48 A0A443RQ67 T1KXG5 A0A1W7R9B1 A0A1B0CDT8 Q16WN2 T1JGA7 A0A1L8DT73 A0A182W515 A0A1L8DT83 A0A182YQG4 A0A2P2HWE1 A0A1W4VJ15 A0A0Q9WWN5 A0A1D2NG26 A0A182R7J8 A0A0Q9WCR2 A0A3B0JWK9 A0A1B0DIL8 A0A0Q9X049 Q29FL9 B4HA29 A0A182PI15 A0A0Q9WNH6 B3NWA6 B3N0B2 A0A0M5J152 M9NFA7 Q9VX91 A0A0P6G4H5 A0A182N0G2 A0A182LSQ1 A0A336LVG5 B4IEW0 A0A1J1HTL0 A0A0P5HK04 A0A0N8AZQ4 A0A1I8MEB9 T1P8G8 A0A0P5LY96 A0A0P5ISW8 A0A0P5FW89
A0A0C9PMM6 A0A2P8Y099 A0A1B6HG01 A0A067QUM0 A0A2J7PG03 A0A232F9W8 K7ISE8 A0A2J7PFZ9 A0A088AH87 A0A2A3ESZ3 E2A1G8 A0A0J7L4E5 E2C439 A0A0M8ZYU1 A0A154P6P7 A0A151K0T7 F4WHK3 A0A158N904 A0A151J379 E9JAU8 A0A1Y1LPA2 A0A151WNN1 A0A1B6DEC1 A0A3L8DSS3 A0A026VTD2 A0A151IBI5 E0VHX5 A0A0L7QWM1 A0A2R7W393 D1ZZM2 A0A146LN83 A0A0A9XCN8 A0A0K8SXL6 A0A1B6EHY3 A0A2H8TIX0 A0A2S2QT54 A0A1B6F6Z8 J9JLI5 N6TQ88 N6ULN5 T1IFX1 N6TMT9 U4UDA6 A0A0P4WDX2 A0A0P4WHX4 A0A087TYA0 A0A224YM26 A0A444SJ90 L7MG75 A0A131YJ48 A0A443RQ67 T1KXG5 A0A1W7R9B1 A0A1B0CDT8 Q16WN2 T1JGA7 A0A1L8DT73 A0A182W515 A0A1L8DT83 A0A182YQG4 A0A2P2HWE1 A0A1W4VJ15 A0A0Q9WWN5 A0A1D2NG26 A0A182R7J8 A0A0Q9WCR2 A0A3B0JWK9 A0A1B0DIL8 A0A0Q9X049 Q29FL9 B4HA29 A0A182PI15 A0A0Q9WNH6 B3NWA6 B3N0B2 A0A0M5J152 M9NFA7 Q9VX91 A0A0P6G4H5 A0A182N0G2 A0A182LSQ1 A0A336LVG5 B4IEW0 A0A1J1HTL0 A0A0P5HK04 A0A0N8AZQ4 A0A1I8MEB9 T1P8G8 A0A0P5LY96 A0A0P5ISW8 A0A0P5FW89
EC Number
2.3.2.27
Pubmed
22118469
28756777
26354079
29403074
24845553
28648823
+ More
20075255 20798317 21719571 21347285 21282665 28004739 30249741 24508170 20566863 18362917 19820115 26823975 25401762 23537049 28797301 25576852 26830274 17510324 25244985 17994087 27289101 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 25315136
20075255 20798317 21719571 21347285 21282665 28004739 30249741 24508170 20566863 18362917 19820115 26823975 25401762 23537049 28797301 25576852 26830274 17510324 25244985 17994087 27289101 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 25315136
EMBL
RSAL01000021
RVE52549.1
AGBW02010576
OWR48249.1
KZ150057
PZC74297.1
+ More
KQ461193 KPJ07005.1 ODYU01002372 SOQ39786.1 GBYB01002374 JAG72141.1 GBYB01002378 JAG72145.1 PYGN01001092 PSN37650.1 GECU01034120 JAS73586.1 KK852918 KDR13847.1 NEVH01025635 PNF15258.1 NNAY01000561 OXU27651.1 PNF15255.1 KZ288189 PBC34634.1 GL435766 EFN72641.1 LBMM01000785 KMQ97501.1 GL452364 EFN77425.1 KQ435794 KOX73890.1 KQ434827 KZC07599.1 KQ981229 KYN44342.1 GL888161 EGI66357.1 ADTU01009011 KQ980290 KYN16918.1 GL770679 EFZ10112.1 GEZM01054746 JAV73426.1 KQ982907 KYQ49453.1 GEDC01027062 GEDC01025510 GEDC01022403 GEDC01014752 GEDC01013257 GEDC01011513 JAS10236.1 JAS11788.1 JAS14895.1 JAS22546.1 JAS24041.1 JAS25785.1 QOIP01000004 RLU23362.1 KK108326 EZA46756.1 KQ978095 KYM97024.1 DS235172 EEB12981.1 KQ414713 KOC62966.1 KK854284 PTY14196.1 KQ971338 EFA01832.1 GDHC01010492 JAQ08137.1 GBHO01025117 JAG18487.1 GBRD01007804 JAG58017.1 GECZ01032241 JAS37528.1 GFXV01002250 MBW14055.1 GGMS01011712 MBY80915.1 GECZ01024126 GECZ01022777 GECZ01014155 JAS45643.1 JAS46992.1 JAS55614.1 ABLF02037916 ABLF02037925 APGK01010906 KB738335 ENN82654.1 APGK01010905 KB738334 ENN82655.1 ACPB03011463 ACPB03011464 ACPB03011465 APGK01029140 APGK01029141 KB740725 ENN79348.1 KB632308 ERL91944.1 GDRN01059226 JAI65518.1 GDRN01059227 GDRN01059225 JAI65517.1 KK117296 KFM70089.1 GFPF01007531 MAA18677.1 SAUD01034488 RXG53467.1 GACK01001833 JAA63201.1 GEDV01010077 JAP78480.1 NCKU01000085 RWS17388.1 CAEY01000695 GFAH01000661 JAV47728.1 AJWK01008222 AJWK01008223 AJWK01008224 AJWK01008225 AJWK01008226 AJWK01008227 AJWK01008228 CH477560 EAT38992.1 JH432199 GFDF01004411 JAV09673.1 GFDF01004412 JAV09672.1 IACF01000279 LAB66072.1 CH933810 KRF94118.1 LJIJ01000051 ODN04223.1 CH940651 KRF82298.1 OUUW01000003 SPP78109.1 AJVK01006142 AJVK01006143 AJVK01006144 AJVK01006145 KRF94117.1 CH379066 EAL31381.2 CH479235 EDW36697.1 KRF82299.1 CH954180 EDV46445.1 CH902640 EDV38316.2 CP012528 ALC49081.1 AE014298 AFH07436.1 AHN59826.1 AY094815 BT044223 GDIQ01038453 JAN56284.1 AXCM01004178 UFQS01000116 UFQT01000116 SSW99947.1 SSX20327.1 CH480832 EDW46214.1 CVRI01000014 CRK89814.1 GDIQ01227028 JAK24697.1 GDIQ01227027 JAK24698.1 KA644937 AFP59566.1 GDIQ01187838 JAK63887.1 GDIQ01209528 JAK42197.1 GDIQ01250824 JAK00901.1
KQ461193 KPJ07005.1 ODYU01002372 SOQ39786.1 GBYB01002374 JAG72141.1 GBYB01002378 JAG72145.1 PYGN01001092 PSN37650.1 GECU01034120 JAS73586.1 KK852918 KDR13847.1 NEVH01025635 PNF15258.1 NNAY01000561 OXU27651.1 PNF15255.1 KZ288189 PBC34634.1 GL435766 EFN72641.1 LBMM01000785 KMQ97501.1 GL452364 EFN77425.1 KQ435794 KOX73890.1 KQ434827 KZC07599.1 KQ981229 KYN44342.1 GL888161 EGI66357.1 ADTU01009011 KQ980290 KYN16918.1 GL770679 EFZ10112.1 GEZM01054746 JAV73426.1 KQ982907 KYQ49453.1 GEDC01027062 GEDC01025510 GEDC01022403 GEDC01014752 GEDC01013257 GEDC01011513 JAS10236.1 JAS11788.1 JAS14895.1 JAS22546.1 JAS24041.1 JAS25785.1 QOIP01000004 RLU23362.1 KK108326 EZA46756.1 KQ978095 KYM97024.1 DS235172 EEB12981.1 KQ414713 KOC62966.1 KK854284 PTY14196.1 KQ971338 EFA01832.1 GDHC01010492 JAQ08137.1 GBHO01025117 JAG18487.1 GBRD01007804 JAG58017.1 GECZ01032241 JAS37528.1 GFXV01002250 MBW14055.1 GGMS01011712 MBY80915.1 GECZ01024126 GECZ01022777 GECZ01014155 JAS45643.1 JAS46992.1 JAS55614.1 ABLF02037916 ABLF02037925 APGK01010906 KB738335 ENN82654.1 APGK01010905 KB738334 ENN82655.1 ACPB03011463 ACPB03011464 ACPB03011465 APGK01029140 APGK01029141 KB740725 ENN79348.1 KB632308 ERL91944.1 GDRN01059226 JAI65518.1 GDRN01059227 GDRN01059225 JAI65517.1 KK117296 KFM70089.1 GFPF01007531 MAA18677.1 SAUD01034488 RXG53467.1 GACK01001833 JAA63201.1 GEDV01010077 JAP78480.1 NCKU01000085 RWS17388.1 CAEY01000695 GFAH01000661 JAV47728.1 AJWK01008222 AJWK01008223 AJWK01008224 AJWK01008225 AJWK01008226 AJWK01008227 AJWK01008228 CH477560 EAT38992.1 JH432199 GFDF01004411 JAV09673.1 GFDF01004412 JAV09672.1 IACF01000279 LAB66072.1 CH933810 KRF94118.1 LJIJ01000051 ODN04223.1 CH940651 KRF82298.1 OUUW01000003 SPP78109.1 AJVK01006142 AJVK01006143 AJVK01006144 AJVK01006145 KRF94117.1 CH379066 EAL31381.2 CH479235 EDW36697.1 KRF82299.1 CH954180 EDV46445.1 CH902640 EDV38316.2 CP012528 ALC49081.1 AE014298 AFH07436.1 AHN59826.1 AY094815 BT044223 GDIQ01038453 JAN56284.1 AXCM01004178 UFQS01000116 UFQT01000116 SSW99947.1 SSX20327.1 CH480832 EDW46214.1 CVRI01000014 CRK89814.1 GDIQ01227028 JAK24697.1 GDIQ01227027 JAK24698.1 KA644937 AFP59566.1 GDIQ01187838 JAK63887.1 GDIQ01209528 JAK42197.1 GDIQ01250824 JAK00901.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000245037
UP000027135
UP000235965
+ More
UP000215335 UP000002358 UP000005203 UP000242457 UP000000311 UP000036403 UP000008237 UP000053105 UP000076502 UP000078541 UP000007755 UP000005205 UP000078492 UP000075809 UP000279307 UP000053097 UP000078542 UP000009046 UP000053825 UP000007266 UP000007819 UP000019118 UP000015103 UP000030742 UP000054359 UP000288706 UP000285301 UP000015104 UP000092461 UP000008820 UP000075920 UP000076408 UP000192221 UP000009192 UP000094527 UP000075900 UP000008792 UP000268350 UP000092462 UP000001819 UP000008744 UP000075885 UP000008711 UP000007801 UP000092553 UP000000803 UP000075884 UP000075883 UP000001292 UP000183832 UP000095301
UP000215335 UP000002358 UP000005203 UP000242457 UP000000311 UP000036403 UP000008237 UP000053105 UP000076502 UP000078541 UP000007755 UP000005205 UP000078492 UP000075809 UP000279307 UP000053097 UP000078542 UP000009046 UP000053825 UP000007266 UP000007819 UP000019118 UP000015103 UP000030742 UP000054359 UP000288706 UP000285301 UP000015104 UP000092461 UP000008820 UP000075920 UP000076408 UP000192221 UP000009192 UP000094527 UP000075900 UP000008792 UP000268350 UP000092462 UP000001819 UP000008744 UP000075885 UP000008711 UP000007801 UP000092553 UP000000803 UP000075884 UP000075883 UP000001292 UP000183832 UP000095301
Interpro
Gene 3D
ProteinModelPortal
A0A3S2LFD2
A0A212F3D3
A0A2W1BLZ8
A0A0N1PGS7
A0A2H1VG67
A0A0C9QG31
+ More
A0A0C9PMM6 A0A2P8Y099 A0A1B6HG01 A0A067QUM0 A0A2J7PG03 A0A232F9W8 K7ISE8 A0A2J7PFZ9 A0A088AH87 A0A2A3ESZ3 E2A1G8 A0A0J7L4E5 E2C439 A0A0M8ZYU1 A0A154P6P7 A0A151K0T7 F4WHK3 A0A158N904 A0A151J379 E9JAU8 A0A1Y1LPA2 A0A151WNN1 A0A1B6DEC1 A0A3L8DSS3 A0A026VTD2 A0A151IBI5 E0VHX5 A0A0L7QWM1 A0A2R7W393 D1ZZM2 A0A146LN83 A0A0A9XCN8 A0A0K8SXL6 A0A1B6EHY3 A0A2H8TIX0 A0A2S2QT54 A0A1B6F6Z8 J9JLI5 N6TQ88 N6ULN5 T1IFX1 N6TMT9 U4UDA6 A0A0P4WDX2 A0A0P4WHX4 A0A087TYA0 A0A224YM26 A0A444SJ90 L7MG75 A0A131YJ48 A0A443RQ67 T1KXG5 A0A1W7R9B1 A0A1B0CDT8 Q16WN2 T1JGA7 A0A1L8DT73 A0A182W515 A0A1L8DT83 A0A182YQG4 A0A2P2HWE1 A0A1W4VJ15 A0A0Q9WWN5 A0A1D2NG26 A0A182R7J8 A0A0Q9WCR2 A0A3B0JWK9 A0A1B0DIL8 A0A0Q9X049 Q29FL9 B4HA29 A0A182PI15 A0A0Q9WNH6 B3NWA6 B3N0B2 A0A0M5J152 M9NFA7 Q9VX91 A0A0P6G4H5 A0A182N0G2 A0A182LSQ1 A0A336LVG5 B4IEW0 A0A1J1HTL0 A0A0P5HK04 A0A0N8AZQ4 A0A1I8MEB9 T1P8G8 A0A0P5LY96 A0A0P5ISW8 A0A0P5FW89
A0A0C9PMM6 A0A2P8Y099 A0A1B6HG01 A0A067QUM0 A0A2J7PG03 A0A232F9W8 K7ISE8 A0A2J7PFZ9 A0A088AH87 A0A2A3ESZ3 E2A1G8 A0A0J7L4E5 E2C439 A0A0M8ZYU1 A0A154P6P7 A0A151K0T7 F4WHK3 A0A158N904 A0A151J379 E9JAU8 A0A1Y1LPA2 A0A151WNN1 A0A1B6DEC1 A0A3L8DSS3 A0A026VTD2 A0A151IBI5 E0VHX5 A0A0L7QWM1 A0A2R7W393 D1ZZM2 A0A146LN83 A0A0A9XCN8 A0A0K8SXL6 A0A1B6EHY3 A0A2H8TIX0 A0A2S2QT54 A0A1B6F6Z8 J9JLI5 N6TQ88 N6ULN5 T1IFX1 N6TMT9 U4UDA6 A0A0P4WDX2 A0A0P4WHX4 A0A087TYA0 A0A224YM26 A0A444SJ90 L7MG75 A0A131YJ48 A0A443RQ67 T1KXG5 A0A1W7R9B1 A0A1B0CDT8 Q16WN2 T1JGA7 A0A1L8DT73 A0A182W515 A0A1L8DT83 A0A182YQG4 A0A2P2HWE1 A0A1W4VJ15 A0A0Q9WWN5 A0A1D2NG26 A0A182R7J8 A0A0Q9WCR2 A0A3B0JWK9 A0A1B0DIL8 A0A0Q9X049 Q29FL9 B4HA29 A0A182PI15 A0A0Q9WNH6 B3NWA6 B3N0B2 A0A0M5J152 M9NFA7 Q9VX91 A0A0P6G4H5 A0A182N0G2 A0A182LSQ1 A0A336LVG5 B4IEW0 A0A1J1HTL0 A0A0P5HK04 A0A0N8AZQ4 A0A1I8MEB9 T1P8G8 A0A0P5LY96 A0A0P5ISW8 A0A0P5FW89
PDB
5TDD
E-value=1.99564e-17,
Score=219
Ontologies
GO
PANTHER
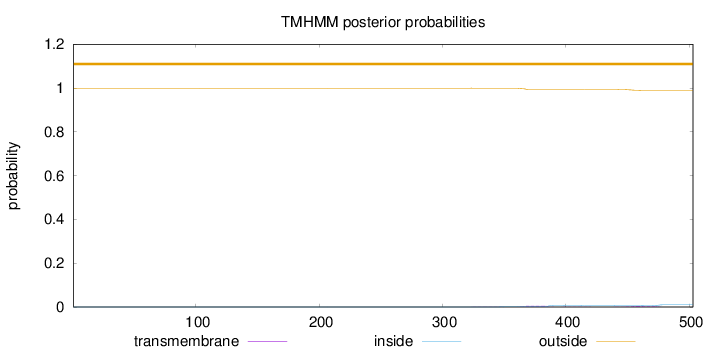
Topology
Length:
503
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23734
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00056
outside
1 - 503
Population Genetic Test Statistics
Pi
216.907658
Theta
158.98475
Tajima's D
1.318708
CLR
0.286409
CSRT
0.744762761861907
Interpretation
Uncertain
