Gene
KWMTBOMO08364 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009282
Annotation
PREDICTED:_protein_kinase_C_and_casein_kinase_substrate_in_neurons_protein_1_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.959
Sequence
CDS
ATGTCGCACCACAGCGACGAGCATGCGTTGCTCACAACCACTTCCGACTCGTTCTGGGAGCCTGGTAACTACAAGAGGACCACCAAGAGAGTAGACGATGGACATCGACTGTGCAGCGAACTTCAAACCCTGGTCCAAGAGCGTGCCGATATCGAGAAAGCGTACGCGAAAAGCCTCAGAGGATGGGCTAAGAAATGGAATGATTTGATCGAAAAGGGACCAGAATACGGCACAATGGAAGCTGCTTGGAAGGGTAGCATGGTGGAAGCGGAACGACTTTCAGACCTCCACCTCAGCGTCAGGGATAGGCTGGTGAACGACGTTATGGCACAGATAAAGAACTGGCAGAAGGAAACTTATCATAAGTCAATGATTCAACTAAAAGAACGCAAGGAAATGGATGAAGCATTCAAGAAGGCCCAGAAACCATGGTCGAAATTACTTCAGAAGGTTGAACGCGCCAGGCTAGAGTACCACACGGCTTGCAAACAAGAAAGAACAGCGCAGAATCAGGAAAGAAACGCCAGCGGAGACAGCTCTTTTAGTCCAGATCAAGTCAAGAAAATGGCAGATCGTGTATCGAAATGTCGCGACGAAGTGTCAAAGAACCGAGAAAAATACCAGACAGCTCTAGCCGAGATCACGGCGTACAACCCTCGATACATAGAGGACATGACTTCAGTATTCGATAGGTGCCAACAGATGGAAGCGCAGCGATTGAAATTCTTCAAAGATGTGCTGTTTAGCTTCCACAAATGCCTCAACATAAGTCAAGAACCGAGCTTACCGCAAATCTACGAAGAGTTCCACCACACGATCAACAACGCAGACAGCCAGAAGGACCTCAAGTGGTGGGCCAACAACCACGGGGTCAATATGGCGATGGCCTGGCCGCAGTTCGAGGAGTACACCGAGGAGTTCCGGGACATCGCCAAGGGCAAGTCCAAGGAGAGCCTGCCCACCGGACCGATTACGTTGCTCAATCAGCGGCCAGTCAGCGAGGATGAGTTACCGCCAATAACGAATAACAAGACGAACAAAAGTAACAATCACTCGGAGCCCCCCGCGCCCGCGCCCGCTGCCCCACCCCTCTCCAACAACACCACCGCCAACAACACCATAGACAAGAAGACTATCAGCGCCCCCATCGCCGTCACTAACGGCACGGCGGGCGGCCCGAAGTCGAACAAGTCGTCGCCCGCGCGGGAGGGCGCCGACAACCCGTTCGAGGAGGAGGAGTGGGACGGGGACGGGGAGGGGGGCGCGCTCACGGACTCCGGCGAGCCCGGCCTGCCCGTGCGCGCGCTCTACGACTACACCGGCGCAGAGAGCGACGAGCTCTCCTTCCGGCAGGGAGACCTGTTTGAGAAGTTGGAGGATGAAGACGAGCAGGGCTGGTGCAAGGGACGCAAGGACGGGCGCGTGGGGCTCTACCCGGCCAACTACGTGGAGCCCGTGGGCCACTAG
Protein
MSHHSDEHALLTTTSDSFWEPGNYKRTTKRVDDGHRLCSELQTLVQERADIEKAYAKSLRGWAKKWNDLIEKGPEYGTMEAAWKGSMVEAERLSDLHLSVRDRLVNDVMAQIKNWQKETYHKSMIQLKERKEMDEAFKKAQKPWSKLLQKVERARLEYHTACKQERTAQNQERNASGDSSFSPDQVKKMADRVSKCRDEVSKNREKYQTALAEITAYNPRYIEDMTSVFDRCQQMEAQRLKFFKDVLFSFHKCLNISQEPSLPQIYEEFHHTINNADSQKDLKWWANNHGVNMAMAWPQFEEYTEEFRDIAKGKSKESLPTGPITLLNQRPVSEDELPPITNNKTNKSNNHSEPPAPAPAAPPLSNNTTANNTIDKKTISAPIAVTNGTAGGPKSNKSSPAREGADNPFEEEEWDGDGEGGALTDSGEPGLPVRALYDYTGAESDELSFRQGDLFEKLEDEDEQGWCKGRKDGRVGLYPANYVEPVGH
Summary
Uniprot
A0A3S2TPS6
A0A1E1WJH9
A0A1E1WPB4
A0A067RQP9
A0A2J7PWV1
K7J511
+ More
A0A2J7PWV9 E2A2F4 A0A2A3EJD8 V9I9X4 A0A195ECI1 F4WQ13 A0A151WKP1 A0A195FH44 A0A026WJF5 A0A1B6EDW3 A0A411G7R9 V9I7R0 A0A195BL78 E9IVU8 A0A023F490 A0A1B6E9A0 A0A151INC5 E0VBJ0 A0A232FJL3 A0A069DZ37 A0A2S2Q3Q6 A0A2H8TK92 A0A2S2NSF1 J9K745 A0A088AC20 A0A154PBA2 A0A0M9A605 A0A3L8E3B8 A0A1L8DME7 A0A310SBP6 A0A1B0C9A8 A0A1L8DM57 A0A0K8TP09 D2A3I3 V5I9J1 A0A1Y1KL32 A0A0A9WGX8 A0A1Q3G3G3 A0A1Q3G3H0 B4PPT6 A0A034V5C8 A0A0R1E0Z7 A0A034V4B7 A0A0L0CDA4 A0A1B0A2F5 D3TNB9 A0A0K8VV18 A0A1A9VF86 A0A084VND4 U5EYM2 B3P319 A0A1A9WFG8 A0A0L7R9L2 B4R1R2 A0A182JDV0 Q9VDI1 A0A0B4KH28 A0A2M4A1J0 W5JBB3 A0A1J1IQK1 A0A2M4BL04 A0A1A9YGX0 A0A1B0BY45 A0A2M3YY04 A0A182V647 T1DHI6 A0A182JTL2 B4II66 B4K8W9 A0A0Q9XDE2 A0A182RC83 A0A0Q9XES1 A0A0Q9X2A9 A0A1D2N7J9 A0A0Q9X483 A0A1I8Q221 A0A182W7A3 W8C0D5 A0A1I8Q210 T1PBU1 A0A182F614 A0A182NU71 A0A1W4UXX2 A0A1B0G369 A0A1W4VB61 A0A226E808 A0A1W4XV85 A0A2J7PWV0 A0A131YZ17 A0A336LU36 A0A182P4V9 A0A182M6A8 A0A1E1X6B9 B3M345
A0A2J7PWV9 E2A2F4 A0A2A3EJD8 V9I9X4 A0A195ECI1 F4WQ13 A0A151WKP1 A0A195FH44 A0A026WJF5 A0A1B6EDW3 A0A411G7R9 V9I7R0 A0A195BL78 E9IVU8 A0A023F490 A0A1B6E9A0 A0A151INC5 E0VBJ0 A0A232FJL3 A0A069DZ37 A0A2S2Q3Q6 A0A2H8TK92 A0A2S2NSF1 J9K745 A0A088AC20 A0A154PBA2 A0A0M9A605 A0A3L8E3B8 A0A1L8DME7 A0A310SBP6 A0A1B0C9A8 A0A1L8DM57 A0A0K8TP09 D2A3I3 V5I9J1 A0A1Y1KL32 A0A0A9WGX8 A0A1Q3G3G3 A0A1Q3G3H0 B4PPT6 A0A034V5C8 A0A0R1E0Z7 A0A034V4B7 A0A0L0CDA4 A0A1B0A2F5 D3TNB9 A0A0K8VV18 A0A1A9VF86 A0A084VND4 U5EYM2 B3P319 A0A1A9WFG8 A0A0L7R9L2 B4R1R2 A0A182JDV0 Q9VDI1 A0A0B4KH28 A0A2M4A1J0 W5JBB3 A0A1J1IQK1 A0A2M4BL04 A0A1A9YGX0 A0A1B0BY45 A0A2M3YY04 A0A182V647 T1DHI6 A0A182JTL2 B4II66 B4K8W9 A0A0Q9XDE2 A0A182RC83 A0A0Q9XES1 A0A0Q9X2A9 A0A1D2N7J9 A0A0Q9X483 A0A1I8Q221 A0A182W7A3 W8C0D5 A0A1I8Q210 T1PBU1 A0A182F614 A0A182NU71 A0A1W4UXX2 A0A1B0G369 A0A1W4VB61 A0A226E808 A0A1W4XV85 A0A2J7PWV0 A0A131YZ17 A0A336LU36 A0A182P4V9 A0A182M6A8 A0A1E1X6B9 B3M345
Pubmed
24845553
20075255
20798317
21719571
24508170
21282665
+ More
25474469 20566863 28648823 26334808 30249741 26369729 18362917 19820115 28004739 25401762 17994087 17550304 25348373 26108605 20353571 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19997509 26109357 26109356 20920257 23761445 27289101 24495485 25315136 26830274 28503490
25474469 20566863 28648823 26334808 30249741 26369729 18362917 19820115 28004739 25401762 17994087 17550304 25348373 26108605 20353571 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19997509 26109357 26109356 20920257 23761445 27289101 24495485 25315136 26830274 28503490
EMBL
RSAL01000028
RVE51835.1
GDQN01003884
JAT87170.1
GDQN01002236
JAT88818.1
+ More
KK852480 KDR22965.1 NEVH01020867 PNF20809.1 PNF20810.1 GL436038 EFN72357.1 KZ288238 PBC31372.1 JR036346 JR036347 AEY57094.1 AEY57095.1 KQ979074 KYN22923.1 GL888262 EGI63711.1 KQ983001 KYQ48449.1 KQ981610 KYN39334.1 KK107168 EZA56120.1 GEDC01001172 JAS36126.1 MH366161 QBB01970.1 JR036342 JR036343 AEY57090.1 AEY57091.1 KQ976441 KYM86422.1 GL766449 EFZ15259.1 GBBI01002396 JAC16316.1 GEDC01002798 JAS34500.1 KQ976940 KYN06969.1 AAZO01000721 DS235030 EEB10746.1 NNAY01000121 OXU30773.1 GBGD01001320 JAC87569.1 GGMS01003146 MBY72349.1 GFXV01002772 MBW14577.1 GGMR01007480 MBY20099.1 ABLF02035160 KQ434847 KZC08470.1 KQ435735 KOX77176.1 QOIP01000001 RLU26885.1 GFDF01006539 JAV07545.1 KQ774368 OAD52324.1 AJWK01002090 AJWK01002091 AJWK01002092 GFDF01006538 JAV07546.1 GDAI01001717 JAI15886.1 KQ971338 EFA01905.1 GALX01002845 JAB65621.1 GEZM01083915 JAV60950.1 GBHO01035907 GBHO01035906 JAG07697.1 JAG07698.1 GFDL01000702 JAV34343.1 GFDL01000701 JAV34344.1 CM000160 EDW96186.1 GAKP01021964 GAKP01021962 JAC36990.1 KRK02972.1 KRK02973.1 GAKP01021960 JAC36992.1 JRES01000558 KNC30245.1 EZ422921 ADD19197.1 GDHF01033603 GDHF01018655 GDHF01009598 GDHF01000574 JAI18711.1 JAI33659.1 JAI42716.1 JAI51740.1 ATLV01014737 KE524984 KFB39478.1 GANO01000264 JAB59607.1 CH954181 EDV48333.1 KQ414624 KOC67557.1 CM000364 EDX12171.1 AE014297 AY069690 AAF55812.2 AAL39835.1 AGB96166.1 AGB96167.1 GGFK01001269 MBW34590.1 ADMH02001879 ETN60693.1 CVRI01000057 CRL02495.1 GGFJ01004502 MBW53643.1 JXJN01022478 GGFM01000398 MBW21149.1 GAMD01002326 JAA99264.1 CH480842 EDW49610.1 CH933806 EDW16566.2 KRG02215.1 KRG02216.1 KRG02214.1 LJIJ01000165 ODN01239.1 KRG02217.1 GAMC01007464 GAMC01007463 JAB99091.1 KA646236 AFP60865.1 CCAG010005574 LNIX01000006 OXA53001.1 PNF20811.1 GEDV01004755 JAP83802.1 UFQT01000003 SSX17158.1 AXCM01003955 GFAC01004602 JAT94586.1 CH902617 EDV42445.1
KK852480 KDR22965.1 NEVH01020867 PNF20809.1 PNF20810.1 GL436038 EFN72357.1 KZ288238 PBC31372.1 JR036346 JR036347 AEY57094.1 AEY57095.1 KQ979074 KYN22923.1 GL888262 EGI63711.1 KQ983001 KYQ48449.1 KQ981610 KYN39334.1 KK107168 EZA56120.1 GEDC01001172 JAS36126.1 MH366161 QBB01970.1 JR036342 JR036343 AEY57090.1 AEY57091.1 KQ976441 KYM86422.1 GL766449 EFZ15259.1 GBBI01002396 JAC16316.1 GEDC01002798 JAS34500.1 KQ976940 KYN06969.1 AAZO01000721 DS235030 EEB10746.1 NNAY01000121 OXU30773.1 GBGD01001320 JAC87569.1 GGMS01003146 MBY72349.1 GFXV01002772 MBW14577.1 GGMR01007480 MBY20099.1 ABLF02035160 KQ434847 KZC08470.1 KQ435735 KOX77176.1 QOIP01000001 RLU26885.1 GFDF01006539 JAV07545.1 KQ774368 OAD52324.1 AJWK01002090 AJWK01002091 AJWK01002092 GFDF01006538 JAV07546.1 GDAI01001717 JAI15886.1 KQ971338 EFA01905.1 GALX01002845 JAB65621.1 GEZM01083915 JAV60950.1 GBHO01035907 GBHO01035906 JAG07697.1 JAG07698.1 GFDL01000702 JAV34343.1 GFDL01000701 JAV34344.1 CM000160 EDW96186.1 GAKP01021964 GAKP01021962 JAC36990.1 KRK02972.1 KRK02973.1 GAKP01021960 JAC36992.1 JRES01000558 KNC30245.1 EZ422921 ADD19197.1 GDHF01033603 GDHF01018655 GDHF01009598 GDHF01000574 JAI18711.1 JAI33659.1 JAI42716.1 JAI51740.1 ATLV01014737 KE524984 KFB39478.1 GANO01000264 JAB59607.1 CH954181 EDV48333.1 KQ414624 KOC67557.1 CM000364 EDX12171.1 AE014297 AY069690 AAF55812.2 AAL39835.1 AGB96166.1 AGB96167.1 GGFK01001269 MBW34590.1 ADMH02001879 ETN60693.1 CVRI01000057 CRL02495.1 GGFJ01004502 MBW53643.1 JXJN01022478 GGFM01000398 MBW21149.1 GAMD01002326 JAA99264.1 CH480842 EDW49610.1 CH933806 EDW16566.2 KRG02215.1 KRG02216.1 KRG02214.1 LJIJ01000165 ODN01239.1 KRG02217.1 GAMC01007464 GAMC01007463 JAB99091.1 KA646236 AFP60865.1 CCAG010005574 LNIX01000006 OXA53001.1 PNF20811.1 GEDV01004755 JAP83802.1 UFQT01000003 SSX17158.1 AXCM01003955 GFAC01004602 JAT94586.1 CH902617 EDV42445.1
Proteomes
UP000283053
UP000027135
UP000235965
UP000002358
UP000000311
UP000242457
+ More
UP000078492 UP000007755 UP000075809 UP000078541 UP000053097 UP000078540 UP000078542 UP000009046 UP000215335 UP000007819 UP000005203 UP000076502 UP000053105 UP000279307 UP000092461 UP000007266 UP000002282 UP000037069 UP000092445 UP000078200 UP000030765 UP000008711 UP000091820 UP000053825 UP000000304 UP000075880 UP000000803 UP000000673 UP000183832 UP000092443 UP000092460 UP000075903 UP000075881 UP000001292 UP000009192 UP000075900 UP000094527 UP000095300 UP000075920 UP000095301 UP000069272 UP000075884 UP000192221 UP000092444 UP000198287 UP000192223 UP000075885 UP000075883 UP000007801
UP000078492 UP000007755 UP000075809 UP000078541 UP000053097 UP000078540 UP000078542 UP000009046 UP000215335 UP000007819 UP000005203 UP000076502 UP000053105 UP000279307 UP000092461 UP000007266 UP000002282 UP000037069 UP000092445 UP000078200 UP000030765 UP000008711 UP000091820 UP000053825 UP000000304 UP000075880 UP000000803 UP000000673 UP000183832 UP000092443 UP000092460 UP000075903 UP000075881 UP000001292 UP000009192 UP000075900 UP000094527 UP000095300 UP000075920 UP000095301 UP000069272 UP000075884 UP000192221 UP000092444 UP000198287 UP000192223 UP000075885 UP000075883 UP000007801
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A3S2TPS6
A0A1E1WJH9
A0A1E1WPB4
A0A067RQP9
A0A2J7PWV1
K7J511
+ More
A0A2J7PWV9 E2A2F4 A0A2A3EJD8 V9I9X4 A0A195ECI1 F4WQ13 A0A151WKP1 A0A195FH44 A0A026WJF5 A0A1B6EDW3 A0A411G7R9 V9I7R0 A0A195BL78 E9IVU8 A0A023F490 A0A1B6E9A0 A0A151INC5 E0VBJ0 A0A232FJL3 A0A069DZ37 A0A2S2Q3Q6 A0A2H8TK92 A0A2S2NSF1 J9K745 A0A088AC20 A0A154PBA2 A0A0M9A605 A0A3L8E3B8 A0A1L8DME7 A0A310SBP6 A0A1B0C9A8 A0A1L8DM57 A0A0K8TP09 D2A3I3 V5I9J1 A0A1Y1KL32 A0A0A9WGX8 A0A1Q3G3G3 A0A1Q3G3H0 B4PPT6 A0A034V5C8 A0A0R1E0Z7 A0A034V4B7 A0A0L0CDA4 A0A1B0A2F5 D3TNB9 A0A0K8VV18 A0A1A9VF86 A0A084VND4 U5EYM2 B3P319 A0A1A9WFG8 A0A0L7R9L2 B4R1R2 A0A182JDV0 Q9VDI1 A0A0B4KH28 A0A2M4A1J0 W5JBB3 A0A1J1IQK1 A0A2M4BL04 A0A1A9YGX0 A0A1B0BY45 A0A2M3YY04 A0A182V647 T1DHI6 A0A182JTL2 B4II66 B4K8W9 A0A0Q9XDE2 A0A182RC83 A0A0Q9XES1 A0A0Q9X2A9 A0A1D2N7J9 A0A0Q9X483 A0A1I8Q221 A0A182W7A3 W8C0D5 A0A1I8Q210 T1PBU1 A0A182F614 A0A182NU71 A0A1W4UXX2 A0A1B0G369 A0A1W4VB61 A0A226E808 A0A1W4XV85 A0A2J7PWV0 A0A131YZ17 A0A336LU36 A0A182P4V9 A0A182M6A8 A0A1E1X6B9 B3M345
A0A2J7PWV9 E2A2F4 A0A2A3EJD8 V9I9X4 A0A195ECI1 F4WQ13 A0A151WKP1 A0A195FH44 A0A026WJF5 A0A1B6EDW3 A0A411G7R9 V9I7R0 A0A195BL78 E9IVU8 A0A023F490 A0A1B6E9A0 A0A151INC5 E0VBJ0 A0A232FJL3 A0A069DZ37 A0A2S2Q3Q6 A0A2H8TK92 A0A2S2NSF1 J9K745 A0A088AC20 A0A154PBA2 A0A0M9A605 A0A3L8E3B8 A0A1L8DME7 A0A310SBP6 A0A1B0C9A8 A0A1L8DM57 A0A0K8TP09 D2A3I3 V5I9J1 A0A1Y1KL32 A0A0A9WGX8 A0A1Q3G3G3 A0A1Q3G3H0 B4PPT6 A0A034V5C8 A0A0R1E0Z7 A0A034V4B7 A0A0L0CDA4 A0A1B0A2F5 D3TNB9 A0A0K8VV18 A0A1A9VF86 A0A084VND4 U5EYM2 B3P319 A0A1A9WFG8 A0A0L7R9L2 B4R1R2 A0A182JDV0 Q9VDI1 A0A0B4KH28 A0A2M4A1J0 W5JBB3 A0A1J1IQK1 A0A2M4BL04 A0A1A9YGX0 A0A1B0BY45 A0A2M3YY04 A0A182V647 T1DHI6 A0A182JTL2 B4II66 B4K8W9 A0A0Q9XDE2 A0A182RC83 A0A0Q9XES1 A0A0Q9X2A9 A0A1D2N7J9 A0A0Q9X483 A0A1I8Q221 A0A182W7A3 W8C0D5 A0A1I8Q210 T1PBU1 A0A182F614 A0A182NU71 A0A1W4UXX2 A0A1B0G369 A0A1W4VB61 A0A226E808 A0A1W4XV85 A0A2J7PWV0 A0A131YZ17 A0A336LU36 A0A182P4V9 A0A182M6A8 A0A1E1X6B9 B3M345
PDB
3I2W
E-value=2.62617e-115,
Score=1063
Ontologies
GO
GO:0016301
GO:0007010
GO:0005543
GO:0005886
GO:0097320
GO:0008092
GO:0005737
GO:0005856
GO:0045211
GO:0000281
GO:0008289
GO:0071212
GO:0007112
GO:0030496
GO:0032154
GO:0045887
GO:0007053
GO:0036090
GO:0061024
GO:0048488
GO:0007269
GO:0016020
GO:0015031
GO:0005515
GO:0003676
GO:0030246
GO:0016853
GO:0019318
GO:0051603
GO:0030145
GO:0006412
GO:0016876
GO:0043039
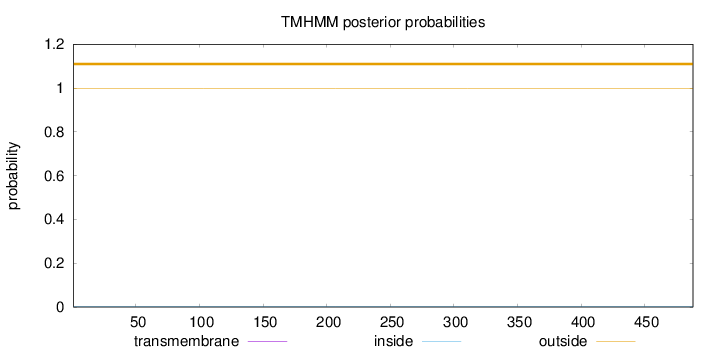
Topology
Length:
488
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00160
outside
1 - 488
Population Genetic Test Statistics
Pi
225.839093
Theta
172.565396
Tajima's D
0.894423
CLR
0.511877
CSRT
0.626968651567422
Interpretation
Uncertain
