Gene
KWMTBOMO08358
Pre Gene Modal
BGIBMGA009230
Annotation
PREDICTED:_cathepsin_O-like_[Amyelois_transitella]
Full name
Tryptophan 2,3-dioxygenase
Alternative Name
Tryptamin 2,3-dioxygenase
Tryptophan oxygenase
Tryptophan pyrrolase
Tryptophanase
Tryptophan oxygenase
Tryptophan pyrrolase
Tryptophanase
Location in the cell
Extracellular Reliability : 1.167
Sequence
CDS
ATGAAAAAATGGTGGAACTGGGTACTCGGTTTTGCTTTGATTTGTTTATTATTCGTAGCTGTTCCTCTTACGTTGTCCAATCAAAAAGGCAAAGAAGAGTTAAAGCCGATGTTTGATGAATATATTCAAAAGTATAATAAAAGTTACAAAAATAAACCCGATGAATACGCAGTGAGATTTGAACATTTTGTGGCATCTGTGCGCGAAATTGAAGAACTAAATGCGGCTGCGCGTGGCCCTGAGGAGCACAGAGCCAGATATGGTTTAACCAGAATCTCGGATATGTCCAAGCTCGAATACAAAGAGATTCACTTATCAGATGAGAGACTTAAGAAATCACCTCATTTGTATGGCAAGAGTTGGAGCAAGTTCAGAGATCAGAAACCTGAAAATACTGTTGGAAATGATGATAATGATACACCCCACAGCAGCCACCAGAGAAATAGAACGCCATGTGCAAATGAACCTCATAATAATGTTTATGTAATAATTAGGAAAAAACGTGCTACTCTTCCATTGAAGGTCGATTGGAGAACGAAAGGAGTTATAGGACCTGTAAGAGATCAGGGTCTATGTGGAGCGTGCTGGGCGTTCAGTACAGTTGGCACTATCGAGTCTATGCAAGCCATCAAGACTGGCACTCTAATGCCACTCAGTGTGCAGGAAGTGATTGACTGCGCCGGTCTAGGCAACAGCGGATGCGCGGGTGGGGACATATGTCTTTTGCTAGACTGGTTGACCATAACCAACACGGCTATACAGCACGAGAAAGAGTACCCGCTCCGTCTCATGAGCGGCGCATGCAAGGCCACTAAGAACGCGACCGGCGTGCGAATATCCACTTTTACTTGTGACGATTTCGTCGGTGCAGAGGAGAAGATATTGGAAGCGTTGGCCACACACGGCCCGGTCACGGTCGCTGTCAACGCCCTAACTTGGCAGAATTATCTCGGCGGAGTCATACAGTATCATTGCAGCGGTGCACCGGCTGACTTGAATCACGCGGTCCAACTTGTTGGCTACGATTTGACGGCCGAAGTGCCTTACTACATCGCCAAGAACTCGTGGGGAGCCGACTACGGAAACGGTGGCTATCTGATGTTGGCCGTGGGCACCAACATTTGTGGTCTGGCCAACGAAGTCGCGACCGTAGACGTTCTATAA
Protein
MKKWWNWVLGFALICLLFVAVPLTLSNQKGKEELKPMFDEYIQKYNKSYKNKPDEYAVRFEHFVASVREIEELNAAARGPEEHRARYGLTRISDMSKLEYKEIHLSDERLKKSPHLYGKSWSKFRDQKPENTVGNDDNDTPHSSHQRNRTPCANEPHNNVYVIIRKKRATLPLKVDWRTKGVIGPVRDQGLCGACWAFSTVGTIESMQAIKTGTLMPLSVQEVIDCAGLGNSGCAGGDICLLLDWLTITNTAIQHEKEYPLRLMSGACKATKNATGVRISTFTCDDFVGAEEKILEALATHGPVTVAVNALTWQNYLGGVIQYHCSGAPADLNHAVQLVGYDLTAEVPYYIAKNSWGADYGNGGYLMLAVGTNICGLANEVATVDVL
Summary
Description
Heme-dependent dioxygenase that catalyzes the oxidative cleavage of the L-tryptophan (L-Trp) pyrrole ring and converts L-tryptophan to N-formyl-L-kynurenine. Catalyzes the oxidative cleavage of the indole moiety.
Catalytic Activity
L-tryptophan + O2 = N-formyl-L-kynurenine
Cofactor
heme
Subunit
Homotetramer. Dimer of dimers.
Similarity
Belongs to the peptidase C1 family.
Belongs to the tryptophan 2,3-dioxygenase family.
Belongs to the tryptophan 2,3-dioxygenase family.
Feature
chain Tryptophan 2,3-dioxygenase
Uniprot
A0A3Q8AYZ4
A0A2H1VBA2
A0A2A4JKQ2
A0A212EPL3
A0A194PY34
A0A437BN63
+ More
J7HI12 A0A2A4IWR3 A0A0N1ICZ1 A0A2W1B435 A0A088AUI0 A0A026W1D4 A0A2A3ERB9 A0A1B6F717 R4WDB3 K7J8W5 A0A0M8ZTL2 E2AWA7 A0A154NZE0 A0A3L8D6J6 E2BJG2 A0A195BSX4 A0A158P1Z3 F4WME2 A0A195F3S1 A0A067R1K0 A0A232EN91 A0A151IL61 A0A195DC55 A0A1V1G5H8 A0A2P8XV78 A0A146LY49 A0A1Y1N308 A0A1B6E1M9 D2A0S2 J3JUX2 E9G115 A0A164LEG2 A0A0P6DUR6 A0A0N8CGR1 A0A0P4VPU0 T1HF18 A0A0P5QE26 A0A1D2MWW1 V5G6Y1 N6UR32 A0A151WQZ2 A0A443SQE0 A0A0N8DME0 A0A0P5DAQ1 N6TT28 B7P7I7 U4U3V2 U6JMC8 A0A0B5J1X2 A0A090XF85 R4G394 A0A1S3DEN6 A0A087V0D1 A0A0P5YPQ9 A0A0P5N3F3 A0A1B6LKV5 A0A226E7F2 A0A0L8FUN9 V4CPT5 A0A1Z5L8G2 A0A182IP04 A0A1S3IVU0 A0A293N5B9 Q17P70 A0A3S1BWC6 A0A0P5G700 A0A1Q3FZ96 A0A182H2Y3 A0A1S3IWG8 A0A2T7PMN4 W5JJ47 A0A0B7ATG4 A0A1I8JW56 Q7QCN0 A0A182IIH7 A0A182LE45 A0A182V6I6 A0A182TTT7 A0A132ACB0 A0A182Y400 A0A3Q3SJ83 A0A1W4ZLU7 W5MNN3 A0A182FQY9 A0A1B0CQA8 A0A2G8KAM0 A0A182VZT9 A0A3Q1CT53 A0A3B4ZKG5
J7HI12 A0A2A4IWR3 A0A0N1ICZ1 A0A2W1B435 A0A088AUI0 A0A026W1D4 A0A2A3ERB9 A0A1B6F717 R4WDB3 K7J8W5 A0A0M8ZTL2 E2AWA7 A0A154NZE0 A0A3L8D6J6 E2BJG2 A0A195BSX4 A0A158P1Z3 F4WME2 A0A195F3S1 A0A067R1K0 A0A232EN91 A0A151IL61 A0A195DC55 A0A1V1G5H8 A0A2P8XV78 A0A146LY49 A0A1Y1N308 A0A1B6E1M9 D2A0S2 J3JUX2 E9G115 A0A164LEG2 A0A0P6DUR6 A0A0N8CGR1 A0A0P4VPU0 T1HF18 A0A0P5QE26 A0A1D2MWW1 V5G6Y1 N6UR32 A0A151WQZ2 A0A443SQE0 A0A0N8DME0 A0A0P5DAQ1 N6TT28 B7P7I7 U4U3V2 U6JMC8 A0A0B5J1X2 A0A090XF85 R4G394 A0A1S3DEN6 A0A087V0D1 A0A0P5YPQ9 A0A0P5N3F3 A0A1B6LKV5 A0A226E7F2 A0A0L8FUN9 V4CPT5 A0A1Z5L8G2 A0A182IP04 A0A1S3IVU0 A0A293N5B9 Q17P70 A0A3S1BWC6 A0A0P5G700 A0A1Q3FZ96 A0A182H2Y3 A0A1S3IWG8 A0A2T7PMN4 W5JJ47 A0A0B7ATG4 A0A1I8JW56 Q7QCN0 A0A182IIH7 A0A182LE45 A0A182V6I6 A0A182TTT7 A0A132ACB0 A0A182Y400 A0A3Q3SJ83 A0A1W4ZLU7 W5MNN3 A0A182FQY9 A0A1B0CQA8 A0A2G8KAM0 A0A182VZT9 A0A3Q1CT53 A0A3B4ZKG5
EC Number
1.13.11.11
Pubmed
22118469
26354079
28756777
24508170
23691247
20075255
+ More
20798317 30249741 21347285 21719571 24845553 28648823 28410430 29403074 26823975 28004739 18362917 19820115 22516182 21292972 27129103 27289101 23537049 25875018 25970599 23254933 28528879 17510324 26483478 20920257 23761445 12364791 14747013 17210077 20966253 26555130 25244985 29023486
20798317 30249741 21347285 21719571 24845553 28648823 28410430 29403074 26823975 28004739 18362917 19820115 22516182 21292972 27129103 27289101 23537049 25875018 25970599 23254933 28528879 17510324 26483478 20920257 23761445 12364791 14747013 17210077 20966253 26555130 25244985 29023486
EMBL
MF155621
ARV79892.1
ODYU01001628
SOQ38119.1
NWSH01001237
PCG72013.1
+ More
AGBW02013432 OWR43445.1 KQ459585 KPI98242.1 RSAL01000028 RVE51843.1 JN400433 AFQ01136.1 NWSH01006263 PCG63553.1 KQ461193 KPJ06987.1 KZ150356 PZC71232.1 KK107488 EZA49885.1 KZ288192 PBC34315.1 GECZ01023730 GECZ01019517 GECZ01007984 JAS46039.1 JAS50252.1 JAS61785.1 AK417518 BAN20733.1 AAZX01013133 AAZX01019176 KQ435850 KOX70875.1 GL443280 EFN62267.1 KQ434778 KZC04434.1 QOIP01000012 RLU15874.1 GL448571 EFN84119.1 KQ976418 KYM89473.1 ADTU01001112 GL888217 EGI64786.1 KQ981855 KYN34827.1 KK852773 KDR16812.1 NNAY01003205 OXU19797.1 KQ977135 KYN05468.1 KQ980989 KYN10495.1 FX985413 BAX07426.1 PYGN01001295 PSN35919.1 GDHC01006146 JAQ12483.1 GEZM01013872 JAV92281.1 GEDC01005466 JAS31832.1 KQ971338 EFA02812.2 BT127037 AEE61999.1 GL732529 EFX86715.1 LRGB01003146 KZS04034.1 GDIQ01073595 JAN21142.1 GDIP01133359 JAL70355.1 GDKW01002189 JAI54406.1 ACPB03015471 GDIQ01122133 JAL29593.1 LJIJ01000446 ODM97411.1 GALX01002621 JAB65845.1 APGK01020083 APGK01020084 APGK01020085 APGK01020086 KB740160 ENN81212.1 KQ982831 KYQ50095.1 NCKV01000807 RWS29685.1 GDIP01019527 JAM84188.1 GDIP01162802 JAJ60600.1 ENN81213.1 ABJB010035484 DS652265 EEC02559.1 KB632077 ERL88569.1 HG710160 CDJ26736.1 KP303286 AJF94884.1 GBIH01000532 JAC94178.1 GAHY01001881 JAA75629.1 KK122590 KFM83070.1 GDIP01056165 JAM47550.1 GDIQ01150141 JAL01585.1 GEBQ01015657 JAT24320.1 LNIX01000006 OXA52877.1 KQ426357 KOF68348.1 KB199753 ESP04425.1 GFJQ02003253 JAW03717.1 GFWV01022862 MAA47589.1 CH477193 EAT48563.1 RQTK01000036 RUS90295.1 GDIQ01245398 JAK06327.1 GFDL01002161 JAV32884.1 JXUM01024214 KQ560683 KXJ81289.1 PZQS01000003 PVD34680.1 ADMH02001265 ETN63323.1 HACG01037454 HACG01037455 CEK84319.1 CEK84320.1 AAAB01008859 EAA08145.5 APCN01000245 JXLN01012453 KPM08509.1 AHAT01015360 AJWK01023339 MRZV01000740 PIK45009.1
AGBW02013432 OWR43445.1 KQ459585 KPI98242.1 RSAL01000028 RVE51843.1 JN400433 AFQ01136.1 NWSH01006263 PCG63553.1 KQ461193 KPJ06987.1 KZ150356 PZC71232.1 KK107488 EZA49885.1 KZ288192 PBC34315.1 GECZ01023730 GECZ01019517 GECZ01007984 JAS46039.1 JAS50252.1 JAS61785.1 AK417518 BAN20733.1 AAZX01013133 AAZX01019176 KQ435850 KOX70875.1 GL443280 EFN62267.1 KQ434778 KZC04434.1 QOIP01000012 RLU15874.1 GL448571 EFN84119.1 KQ976418 KYM89473.1 ADTU01001112 GL888217 EGI64786.1 KQ981855 KYN34827.1 KK852773 KDR16812.1 NNAY01003205 OXU19797.1 KQ977135 KYN05468.1 KQ980989 KYN10495.1 FX985413 BAX07426.1 PYGN01001295 PSN35919.1 GDHC01006146 JAQ12483.1 GEZM01013872 JAV92281.1 GEDC01005466 JAS31832.1 KQ971338 EFA02812.2 BT127037 AEE61999.1 GL732529 EFX86715.1 LRGB01003146 KZS04034.1 GDIQ01073595 JAN21142.1 GDIP01133359 JAL70355.1 GDKW01002189 JAI54406.1 ACPB03015471 GDIQ01122133 JAL29593.1 LJIJ01000446 ODM97411.1 GALX01002621 JAB65845.1 APGK01020083 APGK01020084 APGK01020085 APGK01020086 KB740160 ENN81212.1 KQ982831 KYQ50095.1 NCKV01000807 RWS29685.1 GDIP01019527 JAM84188.1 GDIP01162802 JAJ60600.1 ENN81213.1 ABJB010035484 DS652265 EEC02559.1 KB632077 ERL88569.1 HG710160 CDJ26736.1 KP303286 AJF94884.1 GBIH01000532 JAC94178.1 GAHY01001881 JAA75629.1 KK122590 KFM83070.1 GDIP01056165 JAM47550.1 GDIQ01150141 JAL01585.1 GEBQ01015657 JAT24320.1 LNIX01000006 OXA52877.1 KQ426357 KOF68348.1 KB199753 ESP04425.1 GFJQ02003253 JAW03717.1 GFWV01022862 MAA47589.1 CH477193 EAT48563.1 RQTK01000036 RUS90295.1 GDIQ01245398 JAK06327.1 GFDL01002161 JAV32884.1 JXUM01024214 KQ560683 KXJ81289.1 PZQS01000003 PVD34680.1 ADMH02001265 ETN63323.1 HACG01037454 HACG01037455 CEK84319.1 CEK84320.1 AAAB01008859 EAA08145.5 APCN01000245 JXLN01012453 KPM08509.1 AHAT01015360 AJWK01023339 MRZV01000740 PIK45009.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000283053
UP000053240
UP000005203
+ More
UP000053097 UP000242457 UP000002358 UP000053105 UP000000311 UP000076502 UP000279307 UP000008237 UP000078540 UP000005205 UP000007755 UP000078541 UP000027135 UP000215335 UP000078542 UP000078492 UP000245037 UP000007266 UP000000305 UP000076858 UP000015103 UP000094527 UP000019118 UP000075809 UP000288716 UP000001555 UP000030742 UP000079169 UP000054359 UP000198287 UP000053454 UP000030746 UP000075880 UP000085678 UP000008820 UP000271974 UP000069940 UP000249989 UP000245119 UP000000673 UP000076407 UP000007062 UP000075840 UP000075882 UP000075903 UP000075902 UP000076408 UP000261640 UP000192224 UP000018468 UP000069272 UP000092461 UP000230750 UP000075920 UP000257160 UP000261400
UP000053097 UP000242457 UP000002358 UP000053105 UP000000311 UP000076502 UP000279307 UP000008237 UP000078540 UP000005205 UP000007755 UP000078541 UP000027135 UP000215335 UP000078542 UP000078492 UP000245037 UP000007266 UP000000305 UP000076858 UP000015103 UP000094527 UP000019118 UP000075809 UP000288716 UP000001555 UP000030742 UP000079169 UP000054359 UP000198287 UP000053454 UP000030746 UP000075880 UP000085678 UP000008820 UP000271974 UP000069940 UP000249989 UP000245119 UP000000673 UP000076407 UP000007062 UP000075840 UP000075882 UP000075903 UP000075902 UP000076408 UP000261640 UP000192224 UP000018468 UP000069272 UP000092461 UP000230750 UP000075920 UP000257160 UP000261400
Pfam
Interpro
IPR025660
Pept_his_AS
+ More
IPR039417 Peptidase_C1A_papain-like
IPR038765 Papain-like_cys_pep_sf
IPR013201 Prot_inhib_I29
IPR000169 Pept_cys_AS
IPR000668 Peptidase_C1A_C
IPR011013 Gal_mutarotase_sf_dom
IPR018052 Ald1_epimerase_CS
IPR008183 Aldose_1/G6P_1-epimerase
IPR014718 GH-type_carb-bd
IPR009019 KH_sf_prok-type
IPR015946 KH_dom-like_a/b
IPR004044 KH_dom_type_2
IPR001351 Ribosomal_S3_C
IPR005703 Ribosomal_S3_euk/arc
IPR036419 Ribosomal_S3_C_sf
IPR009071 HMG_box_dom
IPR030089 BAF57
IPR036910 HMG_box_dom_sf
IPR025661 Pept_asp_AS
IPR037217 Trp/Indoleamine_2_3_dOase-like
IPR004981 Trp_2_3_dOase
IPR039417 Peptidase_C1A_papain-like
IPR038765 Papain-like_cys_pep_sf
IPR013201 Prot_inhib_I29
IPR000169 Pept_cys_AS
IPR000668 Peptidase_C1A_C
IPR011013 Gal_mutarotase_sf_dom
IPR018052 Ald1_epimerase_CS
IPR008183 Aldose_1/G6P_1-epimerase
IPR014718 GH-type_carb-bd
IPR009019 KH_sf_prok-type
IPR015946 KH_dom-like_a/b
IPR004044 KH_dom_type_2
IPR001351 Ribosomal_S3_C
IPR005703 Ribosomal_S3_euk/arc
IPR036419 Ribosomal_S3_C_sf
IPR009071 HMG_box_dom
IPR030089 BAF57
IPR036910 HMG_box_dom_sf
IPR025661 Pept_asp_AS
IPR037217 Trp/Indoleamine_2_3_dOase-like
IPR004981 Trp_2_3_dOase
SUPFAM
Gene 3D
ProteinModelPortal
A0A3Q8AYZ4
A0A2H1VBA2
A0A2A4JKQ2
A0A212EPL3
A0A194PY34
A0A437BN63
+ More
J7HI12 A0A2A4IWR3 A0A0N1ICZ1 A0A2W1B435 A0A088AUI0 A0A026W1D4 A0A2A3ERB9 A0A1B6F717 R4WDB3 K7J8W5 A0A0M8ZTL2 E2AWA7 A0A154NZE0 A0A3L8D6J6 E2BJG2 A0A195BSX4 A0A158P1Z3 F4WME2 A0A195F3S1 A0A067R1K0 A0A232EN91 A0A151IL61 A0A195DC55 A0A1V1G5H8 A0A2P8XV78 A0A146LY49 A0A1Y1N308 A0A1B6E1M9 D2A0S2 J3JUX2 E9G115 A0A164LEG2 A0A0P6DUR6 A0A0N8CGR1 A0A0P4VPU0 T1HF18 A0A0P5QE26 A0A1D2MWW1 V5G6Y1 N6UR32 A0A151WQZ2 A0A443SQE0 A0A0N8DME0 A0A0P5DAQ1 N6TT28 B7P7I7 U4U3V2 U6JMC8 A0A0B5J1X2 A0A090XF85 R4G394 A0A1S3DEN6 A0A087V0D1 A0A0P5YPQ9 A0A0P5N3F3 A0A1B6LKV5 A0A226E7F2 A0A0L8FUN9 V4CPT5 A0A1Z5L8G2 A0A182IP04 A0A1S3IVU0 A0A293N5B9 Q17P70 A0A3S1BWC6 A0A0P5G700 A0A1Q3FZ96 A0A182H2Y3 A0A1S3IWG8 A0A2T7PMN4 W5JJ47 A0A0B7ATG4 A0A1I8JW56 Q7QCN0 A0A182IIH7 A0A182LE45 A0A182V6I6 A0A182TTT7 A0A132ACB0 A0A182Y400 A0A3Q3SJ83 A0A1W4ZLU7 W5MNN3 A0A182FQY9 A0A1B0CQA8 A0A2G8KAM0 A0A182VZT9 A0A3Q1CT53 A0A3B4ZKG5
J7HI12 A0A2A4IWR3 A0A0N1ICZ1 A0A2W1B435 A0A088AUI0 A0A026W1D4 A0A2A3ERB9 A0A1B6F717 R4WDB3 K7J8W5 A0A0M8ZTL2 E2AWA7 A0A154NZE0 A0A3L8D6J6 E2BJG2 A0A195BSX4 A0A158P1Z3 F4WME2 A0A195F3S1 A0A067R1K0 A0A232EN91 A0A151IL61 A0A195DC55 A0A1V1G5H8 A0A2P8XV78 A0A146LY49 A0A1Y1N308 A0A1B6E1M9 D2A0S2 J3JUX2 E9G115 A0A164LEG2 A0A0P6DUR6 A0A0N8CGR1 A0A0P4VPU0 T1HF18 A0A0P5QE26 A0A1D2MWW1 V5G6Y1 N6UR32 A0A151WQZ2 A0A443SQE0 A0A0N8DME0 A0A0P5DAQ1 N6TT28 B7P7I7 U4U3V2 U6JMC8 A0A0B5J1X2 A0A090XF85 R4G394 A0A1S3DEN6 A0A087V0D1 A0A0P5YPQ9 A0A0P5N3F3 A0A1B6LKV5 A0A226E7F2 A0A0L8FUN9 V4CPT5 A0A1Z5L8G2 A0A182IP04 A0A1S3IVU0 A0A293N5B9 Q17P70 A0A3S1BWC6 A0A0P5G700 A0A1Q3FZ96 A0A182H2Y3 A0A1S3IWG8 A0A2T7PMN4 W5JJ47 A0A0B7ATG4 A0A1I8JW56 Q7QCN0 A0A182IIH7 A0A182LE45 A0A182V6I6 A0A182TTT7 A0A132ACB0 A0A182Y400 A0A3Q3SJ83 A0A1W4ZLU7 W5MNN3 A0A182FQY9 A0A1B0CQA8 A0A2G8KAM0 A0A182VZT9 A0A3Q1CT53 A0A3B4ZKG5
PDB
6CZK
E-value=1.57687e-35,
Score=374
Ontologies
GO
Topology
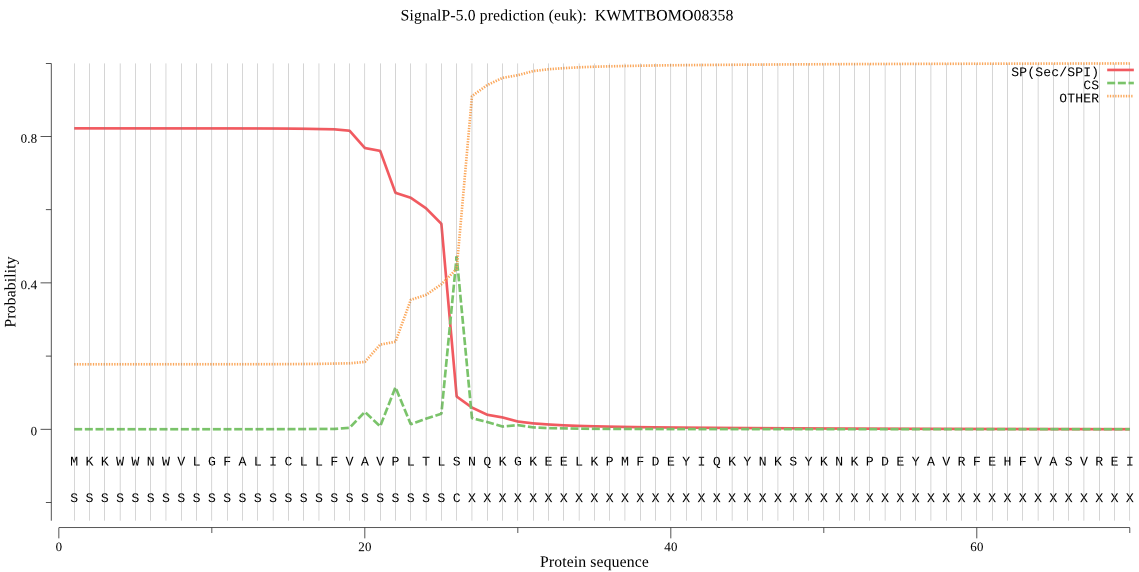
SignalP
Position: 1 - 26,
Likelihood: 0.822540
Length:
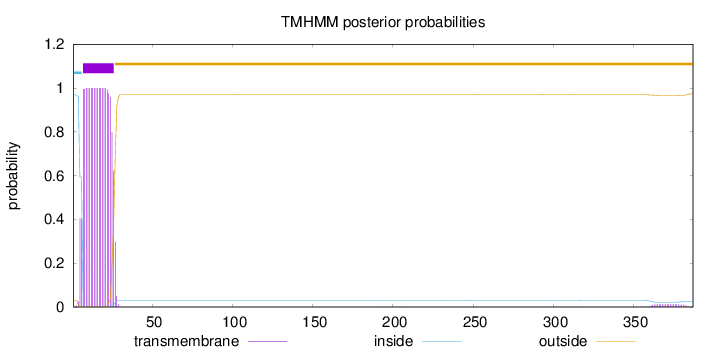
387
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.81586
Exp number, first 60 AAs:
20.53788
Total prob of N-in:
0.97016
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 387
Population Genetic Test Statistics
Pi
198.54699
Theta
176.176307
Tajima's D
0.50459
CLR
0.151759
CSRT
0.515474226288686
Interpretation
Uncertain
