Gene
KWMTBOMO08357
Pre Gene Modal
BGIBMGA009231
Annotation
PREDICTED:_cathepsin_O-like_[Bombyx_mori]
Full name
Cathepsin O
Location in the cell
Extracellular Reliability : 2.011
Sequence
CDS
ATGATCGGACCTGTTTTGAATCAAGGAAATTGCAATGGCTGTTGGGCATACAGCATAGTTGAAACCATGGAGTCTATGGCTGTAAAACAGAAAATGAGTCACACACGTCTTAGCATACAGGAATTACTAGACTGCGTTAGTGTCTGTGAGGGCTGTAATAAAGGCCACGTCGAGGAGGCTCTGAATGAAATGTGTATTAATGATACCGTGGTTACAACAGAAGATAGCTATCCTCTTGGGATAAGTCGTGGAAATTGCAACTCCAGTACATTTAAGGGCAGAAAGATAAAGGAGTACTGGCATTACAGTTTTATGGATGAAAATATTTTACGTCGCGTGATAGCCCACCATGGACCCGTCATAGTCAAGGTGAACGCTCAGCCCTGGATAAATTATCTTGGAGGCATCATCCAGCGCGGTTGTATGCGCAGTGAAGACGGAGCCACTTTGGATCACGCCGTTCAAATAGTCGGCTACAACGCGCGGGGCAGTGTACCCTACTATATAGTGCGCAATTCTTACGGTGCCGACTTCGGTATCGGGGGCTATGTAAAGCTCGCCATGTACCGCAACATTTGCGGTGTAGCTGATGATGTGGCATTTTTGACAGTTTATTAA
Protein
MIGPVLNQGNCNGCWAYSIVETMESMAVKQKMSHTRLSIQELLDCVSVCEGCNKGHVEEALNEMCINDTVVTTEDSYPLGISRGNCNSSTFKGRKIKEYWHYSFMDENILRRVIAHHGPVIVKVNAQPWINYLGGIIQRGCMRSEDGATLDHAVQIVGYNARGSVPYYIVRNSYGADFGIGGYVKLAMYRNICGVADDVAFLTVY
Summary
Catalytic Activity
The recombinant human enzyme hydrolyzes synthetic endopeptidase substrates including Z-Phe-Arg-NHMec and Z-Arg-Arg-NHMec.
Similarity
Belongs to the peptidase C1 family.
Keywords
Complete proteome
Disulfide bond
Glycoprotein
Hydrolase
Lysosome
Protease
Reference proteome
Signal
Thiol protease
Zymogen
Feature
propeptide Activation peptide
chain Cathepsin O
chain Cathepsin O
Uniprot
J7HLR8
A0A2W1B8K7
A0A2H1VBA2
H9JI81
J7HI12
A0A437BN63
+ More
A0A2W1B435 A0A2A4IWR3 A0A2A4JKQ2 A0A2C9JPU1 A0A194PY34 U4U3V2 J3JUX2 N6TT28 N6UR32 A0A212EPL3 A0A3Q8AYZ4 F4WME2 A0A195BSX4 U6JMC8 A0A3S1BWC6 A0A158P1Z3 A0A0P5H6D4 A0A401SW64 A0A2R8Q530 A0A3S3PK68 E7FA09 A0A443SQE0 D2A0S2 V9KZ33 A0A210QAG9 A0A164LEG2 A0A0N8CGR1 A0A0P6DUR6 A0A026W1D4 A0A2R8QDZ6 A0A3B4CWW4 A0A0L8FUN9 A0A0N8DME0 A0A0P5QE26 A0A0P5YPQ9 A0A2T7PMN4 A0A0M8ZTL2 A0A195F3S1 A0A195DC55 A0A2A3ERB9 A0A369SJC0 B3RLZ9 R4WDB3 A0A0B7ATW9 E2AWA7 E9G115 A0A0P5N3F3 A0A088AUI0 A0A151IL61 S4RGW1 V4CPT5 A0A1Y3APM6 A0A0B7ATG4 A0A132ACB0 A0A3L8D6J6 A0A154NZE0 A0A146LY49 W5MNN3 A0A2Y9RK83 A0A1W4ZLU7 A0A293N5B9 A0A401NWW8 A0A0P5DAQ1 G3T124 A0A1S3AHR5 A0A1B0CQA8 W5P9W8 A0A3N0YZV5 K7J8W5 H0WPN5 A0A0B5J1X2 A0A384AA78 G7P6G8 A0A2K6BWC8 A0A3Q7P8N8 A0A2K5U4I7 A0A2K5L851 A0A2K5JKR5 F6RB51 A0A096NQQ3 F1PGK4 G7MS73 A0A452EP08 A0A2K6PMD3 A0A2K6EEJ3 W4ZI04 P43234 A0A287AQD3 A0A0D9S1Q7 E2BJG2 A0A3Q7T5Y0 A0A067R1K0 E1BPI9 A0A341BI86
A0A2W1B435 A0A2A4IWR3 A0A2A4JKQ2 A0A2C9JPU1 A0A194PY34 U4U3V2 J3JUX2 N6TT28 N6UR32 A0A212EPL3 A0A3Q8AYZ4 F4WME2 A0A195BSX4 U6JMC8 A0A3S1BWC6 A0A158P1Z3 A0A0P5H6D4 A0A401SW64 A0A2R8Q530 A0A3S3PK68 E7FA09 A0A443SQE0 D2A0S2 V9KZ33 A0A210QAG9 A0A164LEG2 A0A0N8CGR1 A0A0P6DUR6 A0A026W1D4 A0A2R8QDZ6 A0A3B4CWW4 A0A0L8FUN9 A0A0N8DME0 A0A0P5QE26 A0A0P5YPQ9 A0A2T7PMN4 A0A0M8ZTL2 A0A195F3S1 A0A195DC55 A0A2A3ERB9 A0A369SJC0 B3RLZ9 R4WDB3 A0A0B7ATW9 E2AWA7 E9G115 A0A0P5N3F3 A0A088AUI0 A0A151IL61 S4RGW1 V4CPT5 A0A1Y3APM6 A0A0B7ATG4 A0A132ACB0 A0A3L8D6J6 A0A154NZE0 A0A146LY49 W5MNN3 A0A2Y9RK83 A0A1W4ZLU7 A0A293N5B9 A0A401NWW8 A0A0P5DAQ1 G3T124 A0A1S3AHR5 A0A1B0CQA8 W5P9W8 A0A3N0YZV5 K7J8W5 H0WPN5 A0A0B5J1X2 A0A384AA78 G7P6G8 A0A2K6BWC8 A0A3Q7P8N8 A0A2K5U4I7 A0A2K5L851 A0A2K5JKR5 F6RB51 A0A096NQQ3 F1PGK4 G7MS73 A0A452EP08 A0A2K6PMD3 A0A2K6EEJ3 W4ZI04 P43234 A0A287AQD3 A0A0D9S1Q7 E2BJG2 A0A3Q7T5Y0 A0A067R1K0 E1BPI9 A0A341BI86
EC Number
3.4.22.42
Pubmed
28756777
19121390
15562597
26354079
23537049
22516182
+ More
22118469 21719571 25875018 21347285 30297745 23594743 18362917 19820115 24402279 28812685 24508170 30042472 18719581 23691247 20798317 21292972 23254933 26555130 30249741 26823975 20809919 20075255 22002653 17431167 16341006 25319552 25362486 7929457 15489334 24845553 19393038
22118469 21719571 25875018 21347285 30297745 23594743 18362917 19820115 24402279 28812685 24508170 30042472 18719581 23691247 20798317 21292972 23254933 26555130 30249741 26823975 20809919 20075255 22002653 17431167 16341006 25319552 25362486 7929457 15489334 24845553 19393038
EMBL
JN400432
AFQ01135.1
KZ150356
PZC71231.1
ODYU01001628
SOQ38119.1
+ More
BABH01022310 JN400433 AFQ01136.1 RSAL01000028 RVE51843.1 PZC71232.1 NWSH01006263 PCG63553.1 NWSH01001237 PCG72013.1 KQ459585 KPI98242.1 KB632077 ERL88569.1 BT127037 AEE61999.1 APGK01020083 APGK01020084 APGK01020085 APGK01020086 KB740160 ENN81213.1 ENN81212.1 AGBW02013432 OWR43445.1 MF155621 ARV79892.1 GL888217 EGI64786.1 KQ976418 KYM89473.1 HG710160 CDJ26736.1 RQTK01000036 RUS90295.1 ADTU01001112 GDIQ01232202 JAK19523.1 BEZZ01000618 GCC34642.1 BX005169 NCKU01001735 RWS11380.1 CR931784 NCKV01000807 RWS29685.1 KQ971338 EFA02812.2 JW871605 AFP04123.1 NEDP02004411 OWF45726.1 LRGB01003146 KZS04034.1 GDIP01133359 JAL70355.1 GDIQ01073595 JAN21142.1 KK107488 EZA49885.1 KQ426357 KOF68348.1 GDIP01019527 JAM84188.1 GDIQ01122133 JAL29593.1 GDIP01056165 JAM47550.1 PZQS01000003 PVD34680.1 KQ435850 KOX70875.1 KQ981855 KYN34827.1 KQ980989 KYN10495.1 KZ288192 PBC34315.1 NOWV01000007 RDD46631.1 DS985241 EDV29612.1 AK417518 BAN20733.1 HACG01037453 HACG01037460 CEK84318.1 CEK84325.1 GL443280 EFN62267.1 GL732529 EFX86715.1 GDIQ01150141 JAL01585.1 KQ977135 KYN05468.1 KB199753 ESP04425.1 MUJZ01066012 OTF70390.1 HACG01037454 HACG01037455 CEK84319.1 CEK84320.1 JXLN01012453 KPM08509.1 QOIP01000012 RLU15874.1 KQ434778 KZC04434.1 GDHC01006146 JAQ12483.1 AHAT01015360 GFWV01022862 MAA47589.1 BFAA01005555 GCB65357.1 GDIP01162802 JAJ60600.1 AJWK01023339 AMGL01038845 AMGL01038846 RJVU01018281 ROL51836.1 AAZX01013133 AAZX01019176 AAQR03182834 AAQR03182835 KP303286 AJF94884.1 CM001280 EHH54036.1 AQIA01056791 JSUE03034811 JSUE03034812 AHZZ02028058 AAEX03010073 JV046275 CM001257 AFI36346.1 EHH26267.1 LWLT01000018 AAGJ04098040 AAGJ04098041 X77383 BC049206 AEMK02000065 AQIB01017599 AQIB01017600 GL448571 EFN84119.1 KK852773 KDR16812.1
BABH01022310 JN400433 AFQ01136.1 RSAL01000028 RVE51843.1 PZC71232.1 NWSH01006263 PCG63553.1 NWSH01001237 PCG72013.1 KQ459585 KPI98242.1 KB632077 ERL88569.1 BT127037 AEE61999.1 APGK01020083 APGK01020084 APGK01020085 APGK01020086 KB740160 ENN81213.1 ENN81212.1 AGBW02013432 OWR43445.1 MF155621 ARV79892.1 GL888217 EGI64786.1 KQ976418 KYM89473.1 HG710160 CDJ26736.1 RQTK01000036 RUS90295.1 ADTU01001112 GDIQ01232202 JAK19523.1 BEZZ01000618 GCC34642.1 BX005169 NCKU01001735 RWS11380.1 CR931784 NCKV01000807 RWS29685.1 KQ971338 EFA02812.2 JW871605 AFP04123.1 NEDP02004411 OWF45726.1 LRGB01003146 KZS04034.1 GDIP01133359 JAL70355.1 GDIQ01073595 JAN21142.1 KK107488 EZA49885.1 KQ426357 KOF68348.1 GDIP01019527 JAM84188.1 GDIQ01122133 JAL29593.1 GDIP01056165 JAM47550.1 PZQS01000003 PVD34680.1 KQ435850 KOX70875.1 KQ981855 KYN34827.1 KQ980989 KYN10495.1 KZ288192 PBC34315.1 NOWV01000007 RDD46631.1 DS985241 EDV29612.1 AK417518 BAN20733.1 HACG01037453 HACG01037460 CEK84318.1 CEK84325.1 GL443280 EFN62267.1 GL732529 EFX86715.1 GDIQ01150141 JAL01585.1 KQ977135 KYN05468.1 KB199753 ESP04425.1 MUJZ01066012 OTF70390.1 HACG01037454 HACG01037455 CEK84319.1 CEK84320.1 JXLN01012453 KPM08509.1 QOIP01000012 RLU15874.1 KQ434778 KZC04434.1 GDHC01006146 JAQ12483.1 AHAT01015360 GFWV01022862 MAA47589.1 BFAA01005555 GCB65357.1 GDIP01162802 JAJ60600.1 AJWK01023339 AMGL01038845 AMGL01038846 RJVU01018281 ROL51836.1 AAZX01013133 AAZX01019176 AAQR03182834 AAQR03182835 KP303286 AJF94884.1 CM001280 EHH54036.1 AQIA01056791 JSUE03034811 JSUE03034812 AHZZ02028058 AAEX03010073 JV046275 CM001257 AFI36346.1 EHH26267.1 LWLT01000018 AAGJ04098040 AAGJ04098041 X77383 BC049206 AEMK02000065 AQIB01017599 AQIB01017600 GL448571 EFN84119.1 KK852773 KDR16812.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000076420
UP000053268
UP000030742
+ More
UP000019118 UP000007151 UP000007755 UP000078540 UP000271974 UP000005205 UP000287033 UP000000437 UP000285301 UP000288716 UP000007266 UP000242188 UP000076858 UP000053097 UP000261440 UP000053454 UP000245119 UP000053105 UP000078541 UP000078492 UP000242457 UP000253843 UP000009022 UP000000311 UP000000305 UP000005203 UP000078542 UP000245300 UP000030746 UP000279307 UP000076502 UP000018468 UP000248480 UP000192224 UP000288216 UP000007646 UP000079721 UP000092461 UP000002356 UP000002358 UP000005225 UP000261681 UP000009130 UP000233120 UP000286641 UP000233100 UP000233060 UP000233080 UP000006718 UP000028761 UP000002254 UP000291000 UP000233200 UP000233160 UP000007110 UP000005640 UP000008227 UP000029965 UP000008237 UP000286640 UP000027135 UP000009136 UP000252040
UP000019118 UP000007151 UP000007755 UP000078540 UP000271974 UP000005205 UP000287033 UP000000437 UP000285301 UP000288716 UP000007266 UP000242188 UP000076858 UP000053097 UP000261440 UP000053454 UP000245119 UP000053105 UP000078541 UP000078492 UP000242457 UP000253843 UP000009022 UP000000311 UP000000305 UP000005203 UP000078542 UP000245300 UP000030746 UP000279307 UP000076502 UP000018468 UP000248480 UP000192224 UP000288216 UP000007646 UP000079721 UP000092461 UP000002356 UP000002358 UP000005225 UP000261681 UP000009130 UP000233120 UP000286641 UP000233100 UP000233060 UP000233080 UP000006718 UP000028761 UP000002254 UP000291000 UP000233200 UP000233160 UP000007110 UP000005640 UP000008227 UP000029965 UP000008237 UP000286640 UP000027135 UP000009136 UP000252040
Pfam
Interpro
IPR000668
Peptidase_C1A_C
+ More
IPR025660 Pept_his_AS
IPR039417 Peptidase_C1A_papain-like
IPR038765 Papain-like_cys_pep_sf
IPR013201 Prot_inhib_I29
IPR000169 Pept_cys_AS
IPR011013 Gal_mutarotase_sf_dom
IPR018052 Ald1_epimerase_CS
IPR008183 Aldose_1/G6P_1-epimerase
IPR014718 GH-type_carb-bd
IPR036910 HMG_box_dom_sf
IPR009071 HMG_box_dom
IPR030089 BAF57
IPR025661 Pept_asp_AS
IPR003137 PA_domain
IPR025660 Pept_his_AS
IPR039417 Peptidase_C1A_papain-like
IPR038765 Papain-like_cys_pep_sf
IPR013201 Prot_inhib_I29
IPR000169 Pept_cys_AS
IPR011013 Gal_mutarotase_sf_dom
IPR018052 Ald1_epimerase_CS
IPR008183 Aldose_1/G6P_1-epimerase
IPR014718 GH-type_carb-bd
IPR036910 HMG_box_dom_sf
IPR009071 HMG_box_dom
IPR030089 BAF57
IPR025661 Pept_asp_AS
IPR003137 PA_domain
Gene 3D
ProteinModelPortal
J7HLR8
A0A2W1B8K7
A0A2H1VBA2
H9JI81
J7HI12
A0A437BN63
+ More
A0A2W1B435 A0A2A4IWR3 A0A2A4JKQ2 A0A2C9JPU1 A0A194PY34 U4U3V2 J3JUX2 N6TT28 N6UR32 A0A212EPL3 A0A3Q8AYZ4 F4WME2 A0A195BSX4 U6JMC8 A0A3S1BWC6 A0A158P1Z3 A0A0P5H6D4 A0A401SW64 A0A2R8Q530 A0A3S3PK68 E7FA09 A0A443SQE0 D2A0S2 V9KZ33 A0A210QAG9 A0A164LEG2 A0A0N8CGR1 A0A0P6DUR6 A0A026W1D4 A0A2R8QDZ6 A0A3B4CWW4 A0A0L8FUN9 A0A0N8DME0 A0A0P5QE26 A0A0P5YPQ9 A0A2T7PMN4 A0A0M8ZTL2 A0A195F3S1 A0A195DC55 A0A2A3ERB9 A0A369SJC0 B3RLZ9 R4WDB3 A0A0B7ATW9 E2AWA7 E9G115 A0A0P5N3F3 A0A088AUI0 A0A151IL61 S4RGW1 V4CPT5 A0A1Y3APM6 A0A0B7ATG4 A0A132ACB0 A0A3L8D6J6 A0A154NZE0 A0A146LY49 W5MNN3 A0A2Y9RK83 A0A1W4ZLU7 A0A293N5B9 A0A401NWW8 A0A0P5DAQ1 G3T124 A0A1S3AHR5 A0A1B0CQA8 W5P9W8 A0A3N0YZV5 K7J8W5 H0WPN5 A0A0B5J1X2 A0A384AA78 G7P6G8 A0A2K6BWC8 A0A3Q7P8N8 A0A2K5U4I7 A0A2K5L851 A0A2K5JKR5 F6RB51 A0A096NQQ3 F1PGK4 G7MS73 A0A452EP08 A0A2K6PMD3 A0A2K6EEJ3 W4ZI04 P43234 A0A287AQD3 A0A0D9S1Q7 E2BJG2 A0A3Q7T5Y0 A0A067R1K0 E1BPI9 A0A341BI86
A0A2W1B435 A0A2A4IWR3 A0A2A4JKQ2 A0A2C9JPU1 A0A194PY34 U4U3V2 J3JUX2 N6TT28 N6UR32 A0A212EPL3 A0A3Q8AYZ4 F4WME2 A0A195BSX4 U6JMC8 A0A3S1BWC6 A0A158P1Z3 A0A0P5H6D4 A0A401SW64 A0A2R8Q530 A0A3S3PK68 E7FA09 A0A443SQE0 D2A0S2 V9KZ33 A0A210QAG9 A0A164LEG2 A0A0N8CGR1 A0A0P6DUR6 A0A026W1D4 A0A2R8QDZ6 A0A3B4CWW4 A0A0L8FUN9 A0A0N8DME0 A0A0P5QE26 A0A0P5YPQ9 A0A2T7PMN4 A0A0M8ZTL2 A0A195F3S1 A0A195DC55 A0A2A3ERB9 A0A369SJC0 B3RLZ9 R4WDB3 A0A0B7ATW9 E2AWA7 E9G115 A0A0P5N3F3 A0A088AUI0 A0A151IL61 S4RGW1 V4CPT5 A0A1Y3APM6 A0A0B7ATG4 A0A132ACB0 A0A3L8D6J6 A0A154NZE0 A0A146LY49 W5MNN3 A0A2Y9RK83 A0A1W4ZLU7 A0A293N5B9 A0A401NWW8 A0A0P5DAQ1 G3T124 A0A1S3AHR5 A0A1B0CQA8 W5P9W8 A0A3N0YZV5 K7J8W5 H0WPN5 A0A0B5J1X2 A0A384AA78 G7P6G8 A0A2K6BWC8 A0A3Q7P8N8 A0A2K5U4I7 A0A2K5L851 A0A2K5JKR5 F6RB51 A0A096NQQ3 F1PGK4 G7MS73 A0A452EP08 A0A2K6PMD3 A0A2K6EEJ3 W4ZI04 P43234 A0A287AQD3 A0A0D9S1Q7 E2BJG2 A0A3Q7T5Y0 A0A067R1K0 E1BPI9 A0A341BI86
PDB
3H7D
E-value=1.15336e-22,
Score=260
Ontologies
GO
PANTHER
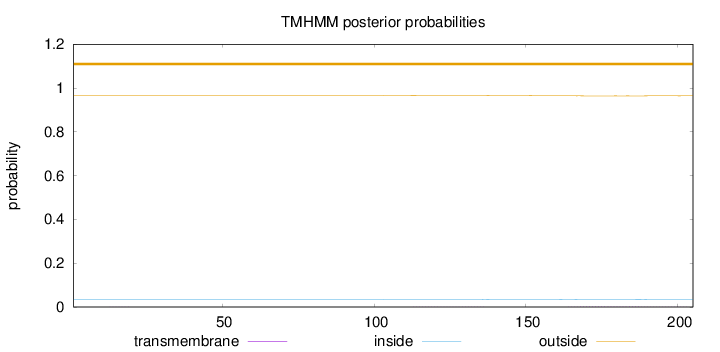
Topology
Subcellular location
Lysosome
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06138
Exp number, first 60 AAs:
0.00223
Total prob of N-in:
0.03425
outside
1 - 205
Population Genetic Test Statistics
Pi
21.560728
Theta
25.318317
Tajima's D
-0.56305
CLR
19.620751
CSRT
0.232988350582471
Interpretation
Uncertain
