Gene
KWMTBOMO08356 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009232
Annotation
PREDICTED:_aldose_1-epimerase_[Papilio_polytes]
Full name
Aldose 1-epimerase
Alternative Name
Galactose mutarotase
Location in the cell
Extracellular Reliability : 1.917
Sequence
CDS
ATGGTGACTTTGAATGTTGAGGATTTTGGACAGCATCATGGAGAGACGGTTTCAAAATTCACATGGAGGGCATCTGATGGCTTCTCAGTGTCCGTCATTTCATATGGGGCTACCATACAGTCAATTCAGGTACCGGACAAATACGGAATCACCAGTGACGTTGTTCTGGGCTTTGACGAACTCGACGGGTACGTGAACAGAAACACACCGTACTTAGGGACCACGGTCGGCCGCTGTGCCAACAGGATCGGCGGAGCAAAATTCAGCATCGACGGCACGACTTATCAACTCGCTAACAACATTGGGAAAGATCATTTACATGGCGGGATTAACGGATTTAATAAGGCGAATTGGAATTCCACCGTTGACGGTACTAAAGTGATATTTAGTTATCTATCAAAGGACGGCGAAGAAGGATACCCGGGTGATCTTATTACTAACATTACTTATGAAGTTACCGAGGACAATGCCTTGCATGTCGACTTTATGTCGACGACAACTAAAAAGACCGTTGTAAATCTGACAAACCATTCGTATTTTAATTTGGCTGGCCACGAAACTGGTGTCCAAGAGATCTACAACCATATCTTTGTTATAAACGCTGACAAGATCACGGAGACGGATTCTGGATCGATTCCGACTGGTGGTTTTATAAGCGTAGGAGGAACGCCCTACGATCTTCGCGTCCCAACTAAACTCGGCGACGTCATAAATAAAACTGGGAACGGTTTCGATGACAACTTCTGCATCTCAACGTACACAAATAAATCATTAAATTTCGTGTCTCGGATCCTTCATCCATCATCGGGACGCACTCTCGAAGTCTACAGCGATCAGCCCGGAGTCCAGTTTTACACCTCTAACTCCCTCCCTGCTCCTCAAGAGTCTGCTCTGGTTGGTAAACAGGGAGTTGGCTACAGACGTCACGGCGCGTTCTGTCTCGAGACCCAAAATTACCCGGACGCTGTCCATCACACGAATTTCCCGAGAGCAGTTCTGTATCCTGGCGAAGTGTATAAACATAAAGTGGTGTACAGATTTGCTGCTGCTGCTGCAGAAAAGCTGCAGTATCCTATTGTTTTGCCTGCGTGA
Protein
MVTLNVEDFGQHHGETVSKFTWRASDGFSVSVISYGATIQSIQVPDKYGITSDVVLGFDELDGYVNRNTPYLGTTVGRCANRIGGAKFSIDGTTYQLANNIGKDHLHGGINGFNKANWNSTVDGTKVIFSYLSKDGEEGYPGDLITNITYEVTEDNALHVDFMSTTTKKTVVNLTNHSYFNLAGHETGVQEIYNHIFVINADKITETDSGSIPTGGFISVGGTPYDLRVPTKLGDVINKTGNGFDDNFCISTYTNKSLNFVSRILHPSSGRTLEVYSDQPGVQFYTSNSLPAPQESALVGKQGVGYRRHGAFCLETQNYPDAVHHTNFPRAVLYPGEVYKHKVVYRFAAAAAEKLQYPIVLPA
Summary
Description
Converts alpha-aldose to the beta-anomer.
Catalytic Activity
alpha-D-glucose = beta-D-glucose
Similarity
Belongs to the aldose epimerase family.
Belongs to the peptidase C1 family.
Belongs to the peptidase C1 family.
Feature
chain Aldose 1-epimerase
Uniprot
H9JI83
A0A3S2NIF1
A0A2W1BEF6
A0A0N1IEM2
A0A2H1VBA2
A0A212EZ93
+ More
A0A2H1V8W8 A0A023ERU2 A0A084WTH2 A0A182GEB2 A0A182GGN4 A0A023ETD0 Q16SI6 A0A182IL09 A0A1Y1MZX9 A0A1S4FQS0 A0A1Y1N199 A0A182VDL1 A0A182KZV5 A0A182YJR4 A0A182W357 A0A182XF42 Q5TT05 A0A182FQE5 A0A182HVL0 A0A1Q3FR36 A0A182RI49 A0A182LTE1 A0A182JW77 A0A2M4BM71 W5JSN6 A0A2M4BLP1 A0A182NCS4 A0A182QY74 A0A1L8E1K3 A0A2M4BNQ6 B0W3B1 A0A1L8E1P9 A0A182SEM9 A0A182TQD7 A0A182PLE7 A0A2M4A711 A0A2M4A685 A0A2M4A6N3 A0A1B0GI99 A0A154PBN4 A0A1J1I063 E9IX12 E2BW64 V5GTG1 K7J742 A0A151I6V8 U4U9P8 A0A139WPN3 A0A194PY24 A0A336MR65 U5EP71 A0A336MNB8 A0A195B0P1 A0A151WS16 A0A195FBH6 A0A151JB89 A0A067R8C0 N6TS18 A0A2P8Y947 F4WWB1 A0A2J7QHX4 A0A3L8D803 A0A0M8ZP90 A0A026X2U9 A0A158NKI4 A0A0L7RDB7 D6WGD2 A0A139WPU0 A0A087ZNJ7 J3JUV0 A0A2J7QHY8 A0A2A3E2I6 N6TV79 E2AC71 A0A336LQN6 B4IZS5 B4LHE1 E0VPV6 C3Z3S8 A0A2M4CNH5 A0A0J7L5T4 A0A3B0KFK6 B4KWQ0 A0A1W4V9H2 A0A1S3H0U3 B4QJ93 A0A0L7K3R6 B4HUP1 A0A1I8PV53 A0A1B0CYR6 C5WLR2 Q8MT28 D3TPX1
A0A2H1V8W8 A0A023ERU2 A0A084WTH2 A0A182GEB2 A0A182GGN4 A0A023ETD0 Q16SI6 A0A182IL09 A0A1Y1MZX9 A0A1S4FQS0 A0A1Y1N199 A0A182VDL1 A0A182KZV5 A0A182YJR4 A0A182W357 A0A182XF42 Q5TT05 A0A182FQE5 A0A182HVL0 A0A1Q3FR36 A0A182RI49 A0A182LTE1 A0A182JW77 A0A2M4BM71 W5JSN6 A0A2M4BLP1 A0A182NCS4 A0A182QY74 A0A1L8E1K3 A0A2M4BNQ6 B0W3B1 A0A1L8E1P9 A0A182SEM9 A0A182TQD7 A0A182PLE7 A0A2M4A711 A0A2M4A685 A0A2M4A6N3 A0A1B0GI99 A0A154PBN4 A0A1J1I063 E9IX12 E2BW64 V5GTG1 K7J742 A0A151I6V8 U4U9P8 A0A139WPN3 A0A194PY24 A0A336MR65 U5EP71 A0A336MNB8 A0A195B0P1 A0A151WS16 A0A195FBH6 A0A151JB89 A0A067R8C0 N6TS18 A0A2P8Y947 F4WWB1 A0A2J7QHX4 A0A3L8D803 A0A0M8ZP90 A0A026X2U9 A0A158NKI4 A0A0L7RDB7 D6WGD2 A0A139WPU0 A0A087ZNJ7 J3JUV0 A0A2J7QHY8 A0A2A3E2I6 N6TV79 E2AC71 A0A336LQN6 B4IZS5 B4LHE1 E0VPV6 C3Z3S8 A0A2M4CNH5 A0A0J7L5T4 A0A3B0KFK6 B4KWQ0 A0A1W4V9H2 A0A1S3H0U3 B4QJ93 A0A0L7K3R6 B4HUP1 A0A1I8PV53 A0A1B0CYR6 C5WLR2 Q8MT28 D3TPX1
EC Number
5.1.3.3
Pubmed
19121390
28756777
26354079
22118469
24945155
24438588
+ More
26483478 17510324 28004739 20966253 25244985 12364791 14747013 17210077 20920257 23761445 21282665 20798317 20075255 23537049 18362917 19820115 24845553 29403074 21719571 30249741 24508170 21347285 22516182 17994087 20566863 18563158 22936249 26227816 20353571
26483478 17510324 28004739 20966253 25244985 12364791 14747013 17210077 20920257 23761445 21282665 20798317 20075255 23537049 18362917 19820115 24845553 29403074 21719571 30249741 24508170 21347285 22516182 17994087 20566863 18563158 22936249 26227816 20353571
EMBL
BABH01022310
RSAL01000028
RVE51845.1
KZ150356
PZC71230.1
KQ461193
+ More
KPJ06986.1 ODYU01001628 SOQ38119.1 AGBW02011349 OWR46800.1 ODYU01001100 SOQ36852.1 GAPW01001690 JAC11908.1 ATLV01026892 KE525420 KFB53516.1 JXUM01057419 KQ561954 KXJ77062.1 JXUM01061998 KQ562178 KXJ76481.1 GAPW01001502 JAC12096.1 CH477673 EAT37419.1 GEZM01016664 JAV91209.1 GEZM01016665 JAV91208.1 AAAB01008898 EAL40526.3 APCN01005799 GFDL01005021 JAV30024.1 AXCM01001801 GGFJ01005004 MBW54145.1 ADMH02000200 ETN67387.1 GGFJ01004859 MBW54000.1 AXCN02001584 GFDF01001474 JAV12610.1 GGFJ01005267 MBW54408.1 DS231831 EDS31254.1 GFDF01001475 JAV12609.1 GGFK01003097 MBW36418.1 GGFK01002968 MBW36289.1 GGFK01003094 MBW36415.1 AJWK01013596 AJWK01013597 KQ434856 KZC08668.1 CVRI01000037 CRK93653.1 GL766616 EFZ14909.1 GL451091 EFN80096.1 GALX01004943 JAB63523.1 AAZX01002970 KQ978456 KYM93890.1 KYM93891.1 KB631843 ERL86660.1 KQ971307 KYB29856.1 KQ459585 KPI98241.1 UFQT01002209 SSX32932.1 GANO01000283 JAB59588.1 UFQT01001726 SSX31540.1 KQ976692 KYM77862.1 KQ982805 KYQ50465.1 KQ981693 KYN37773.1 KQ979182 KYN22340.1 KK852869 KDR14666.1 APGK01004458 APGK01004459 KB735765 ENN83304.1 PYGN01000790 PSN40786.1 GL888406 EGI61453.1 NEVH01013964 PNF28188.1 QOIP01000011 RLU16585.1 KQ438551 KOX67206.1 KK107021 EZA62346.1 ADTU01018854 KQ414614 KOC68795.1 KQ971329 EFA01124.1 KYB29855.1 BT127015 AEE61977.1 PNF28186.1 KZ288427 PBC25910.1 APGK01000121 KB741373 ENN70182.1 GL438428 EFN68965.1 UFQT01000108 SSX20115.1 CH916366 EDV95660.1 CH940647 EDW68471.1 DS235379 EEB15412.1 GG666577 EEN52842.1 GGFL01002702 MBW66880.1 LBMM01000640 KMQ97918.1 OUUW01000009 SPP85109.1 CH933809 EDW17497.1 CM000363 CM002912 EDX09402.1 KMY97861.1 JTDY01012359 KOB52288.1 CH480817 EDW50662.1 AJVK01020327 BT088877 ACS92849.1 AY118414 AAM48443.1 EZ423473 ADD19749.1
KPJ06986.1 ODYU01001628 SOQ38119.1 AGBW02011349 OWR46800.1 ODYU01001100 SOQ36852.1 GAPW01001690 JAC11908.1 ATLV01026892 KE525420 KFB53516.1 JXUM01057419 KQ561954 KXJ77062.1 JXUM01061998 KQ562178 KXJ76481.1 GAPW01001502 JAC12096.1 CH477673 EAT37419.1 GEZM01016664 JAV91209.1 GEZM01016665 JAV91208.1 AAAB01008898 EAL40526.3 APCN01005799 GFDL01005021 JAV30024.1 AXCM01001801 GGFJ01005004 MBW54145.1 ADMH02000200 ETN67387.1 GGFJ01004859 MBW54000.1 AXCN02001584 GFDF01001474 JAV12610.1 GGFJ01005267 MBW54408.1 DS231831 EDS31254.1 GFDF01001475 JAV12609.1 GGFK01003097 MBW36418.1 GGFK01002968 MBW36289.1 GGFK01003094 MBW36415.1 AJWK01013596 AJWK01013597 KQ434856 KZC08668.1 CVRI01000037 CRK93653.1 GL766616 EFZ14909.1 GL451091 EFN80096.1 GALX01004943 JAB63523.1 AAZX01002970 KQ978456 KYM93890.1 KYM93891.1 KB631843 ERL86660.1 KQ971307 KYB29856.1 KQ459585 KPI98241.1 UFQT01002209 SSX32932.1 GANO01000283 JAB59588.1 UFQT01001726 SSX31540.1 KQ976692 KYM77862.1 KQ982805 KYQ50465.1 KQ981693 KYN37773.1 KQ979182 KYN22340.1 KK852869 KDR14666.1 APGK01004458 APGK01004459 KB735765 ENN83304.1 PYGN01000790 PSN40786.1 GL888406 EGI61453.1 NEVH01013964 PNF28188.1 QOIP01000011 RLU16585.1 KQ438551 KOX67206.1 KK107021 EZA62346.1 ADTU01018854 KQ414614 KOC68795.1 KQ971329 EFA01124.1 KYB29855.1 BT127015 AEE61977.1 PNF28186.1 KZ288427 PBC25910.1 APGK01000121 KB741373 ENN70182.1 GL438428 EFN68965.1 UFQT01000108 SSX20115.1 CH916366 EDV95660.1 CH940647 EDW68471.1 DS235379 EEB15412.1 GG666577 EEN52842.1 GGFL01002702 MBW66880.1 LBMM01000640 KMQ97918.1 OUUW01000009 SPP85109.1 CH933809 EDW17497.1 CM000363 CM002912 EDX09402.1 KMY97861.1 JTDY01012359 KOB52288.1 CH480817 EDW50662.1 AJVK01020327 BT088877 ACS92849.1 AY118414 AAM48443.1 EZ423473 ADD19749.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000030765
UP000069940
+ More
UP000249989 UP000008820 UP000075880 UP000075903 UP000075882 UP000076408 UP000075920 UP000076407 UP000007062 UP000069272 UP000075840 UP000075900 UP000075883 UP000075881 UP000000673 UP000075884 UP000075886 UP000002320 UP000075901 UP000075902 UP000075885 UP000092461 UP000076502 UP000183832 UP000008237 UP000002358 UP000078542 UP000030742 UP000007266 UP000053268 UP000078540 UP000075809 UP000078541 UP000078492 UP000027135 UP000019118 UP000245037 UP000007755 UP000235965 UP000279307 UP000053105 UP000053097 UP000005205 UP000053825 UP000005203 UP000242457 UP000000311 UP000001070 UP000008792 UP000009046 UP000001554 UP000036403 UP000268350 UP000009192 UP000192221 UP000085678 UP000000304 UP000037510 UP000001292 UP000095300 UP000092462
UP000249989 UP000008820 UP000075880 UP000075903 UP000075882 UP000076408 UP000075920 UP000076407 UP000007062 UP000069272 UP000075840 UP000075900 UP000075883 UP000075881 UP000000673 UP000075884 UP000075886 UP000002320 UP000075901 UP000075902 UP000075885 UP000092461 UP000076502 UP000183832 UP000008237 UP000002358 UP000078542 UP000030742 UP000007266 UP000053268 UP000078540 UP000075809 UP000078541 UP000078492 UP000027135 UP000019118 UP000245037 UP000007755 UP000235965 UP000279307 UP000053105 UP000053097 UP000005205 UP000053825 UP000005203 UP000242457 UP000000311 UP000001070 UP000008792 UP000009046 UP000001554 UP000036403 UP000268350 UP000009192 UP000192221 UP000085678 UP000000304 UP000037510 UP000001292 UP000095300 UP000092462
Interpro
IPR015443
Aldose_1-epimerase
+ More
IPR014718 GH-type_carb-bd
IPR011013 Gal_mutarotase_sf_dom
IPR018052 Ald1_epimerase_CS
IPR008183 Aldose_1/G6P_1-epimerase
IPR000169 Pept_cys_AS
IPR000668 Peptidase_C1A_C
IPR039417 Peptidase_C1A_papain-like
IPR038765 Papain-like_cys_pep_sf
IPR025660 Pept_his_AS
IPR013201 Prot_inhib_I29
IPR014718 GH-type_carb-bd
IPR011013 Gal_mutarotase_sf_dom
IPR018052 Ald1_epimerase_CS
IPR008183 Aldose_1/G6P_1-epimerase
IPR000169 Pept_cys_AS
IPR000668 Peptidase_C1A_C
IPR039417 Peptidase_C1A_papain-like
IPR038765 Papain-like_cys_pep_sf
IPR025660 Pept_his_AS
IPR013201 Prot_inhib_I29
Gene 3D
ProteinModelPortal
H9JI83
A0A3S2NIF1
A0A2W1BEF6
A0A0N1IEM2
A0A2H1VBA2
A0A212EZ93
+ More
A0A2H1V8W8 A0A023ERU2 A0A084WTH2 A0A182GEB2 A0A182GGN4 A0A023ETD0 Q16SI6 A0A182IL09 A0A1Y1MZX9 A0A1S4FQS0 A0A1Y1N199 A0A182VDL1 A0A182KZV5 A0A182YJR4 A0A182W357 A0A182XF42 Q5TT05 A0A182FQE5 A0A182HVL0 A0A1Q3FR36 A0A182RI49 A0A182LTE1 A0A182JW77 A0A2M4BM71 W5JSN6 A0A2M4BLP1 A0A182NCS4 A0A182QY74 A0A1L8E1K3 A0A2M4BNQ6 B0W3B1 A0A1L8E1P9 A0A182SEM9 A0A182TQD7 A0A182PLE7 A0A2M4A711 A0A2M4A685 A0A2M4A6N3 A0A1B0GI99 A0A154PBN4 A0A1J1I063 E9IX12 E2BW64 V5GTG1 K7J742 A0A151I6V8 U4U9P8 A0A139WPN3 A0A194PY24 A0A336MR65 U5EP71 A0A336MNB8 A0A195B0P1 A0A151WS16 A0A195FBH6 A0A151JB89 A0A067R8C0 N6TS18 A0A2P8Y947 F4WWB1 A0A2J7QHX4 A0A3L8D803 A0A0M8ZP90 A0A026X2U9 A0A158NKI4 A0A0L7RDB7 D6WGD2 A0A139WPU0 A0A087ZNJ7 J3JUV0 A0A2J7QHY8 A0A2A3E2I6 N6TV79 E2AC71 A0A336LQN6 B4IZS5 B4LHE1 E0VPV6 C3Z3S8 A0A2M4CNH5 A0A0J7L5T4 A0A3B0KFK6 B4KWQ0 A0A1W4V9H2 A0A1S3H0U3 B4QJ93 A0A0L7K3R6 B4HUP1 A0A1I8PV53 A0A1B0CYR6 C5WLR2 Q8MT28 D3TPX1
A0A2H1V8W8 A0A023ERU2 A0A084WTH2 A0A182GEB2 A0A182GGN4 A0A023ETD0 Q16SI6 A0A182IL09 A0A1Y1MZX9 A0A1S4FQS0 A0A1Y1N199 A0A182VDL1 A0A182KZV5 A0A182YJR4 A0A182W357 A0A182XF42 Q5TT05 A0A182FQE5 A0A182HVL0 A0A1Q3FR36 A0A182RI49 A0A182LTE1 A0A182JW77 A0A2M4BM71 W5JSN6 A0A2M4BLP1 A0A182NCS4 A0A182QY74 A0A1L8E1K3 A0A2M4BNQ6 B0W3B1 A0A1L8E1P9 A0A182SEM9 A0A182TQD7 A0A182PLE7 A0A2M4A711 A0A2M4A685 A0A2M4A6N3 A0A1B0GI99 A0A154PBN4 A0A1J1I063 E9IX12 E2BW64 V5GTG1 K7J742 A0A151I6V8 U4U9P8 A0A139WPN3 A0A194PY24 A0A336MR65 U5EP71 A0A336MNB8 A0A195B0P1 A0A151WS16 A0A195FBH6 A0A151JB89 A0A067R8C0 N6TS18 A0A2P8Y947 F4WWB1 A0A2J7QHX4 A0A3L8D803 A0A0M8ZP90 A0A026X2U9 A0A158NKI4 A0A0L7RDB7 D6WGD2 A0A139WPU0 A0A087ZNJ7 J3JUV0 A0A2J7QHY8 A0A2A3E2I6 N6TV79 E2AC71 A0A336LQN6 B4IZS5 B4LHE1 E0VPV6 C3Z3S8 A0A2M4CNH5 A0A0J7L5T4 A0A3B0KFK6 B4KWQ0 A0A1W4V9H2 A0A1S3H0U3 B4QJ93 A0A0L7K3R6 B4HUP1 A0A1I8PV53 A0A1B0CYR6 C5WLR2 Q8MT28 D3TPX1
PDB
1SO0
E-value=3.17615e-70,
Score=673
Ontologies
PATHWAY
GO
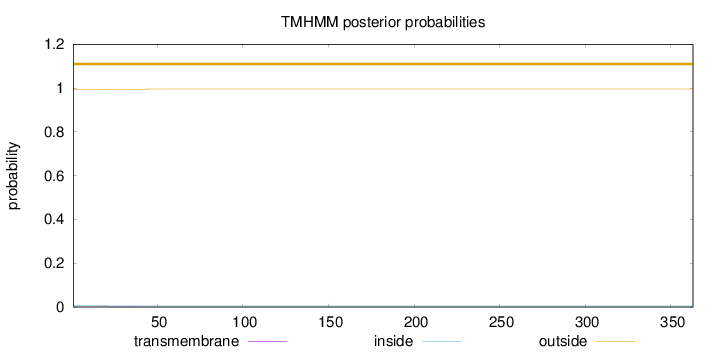
Topology
Length:
363
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02724
Exp number, first 60 AAs:
0.02675
Total prob of N-in:
0.00566
outside
1 - 363
Population Genetic Test Statistics
Pi
163.544111
Theta
19.802176
Tajima's D
-0.855667
CLR
2.949059
CSRT
0.168641567921604
Interpretation
Uncertain
