Pre Gene Modal
BGIBMGA009276
Annotation
PREDICTED:_tryptophan_2?3-dioxygenase_isoform_X2_[Bombyx_mori]
Full name
Tryptophan 2,3-dioxygenase
Alternative Name
Tryptamin 2,3-dioxygenase
Tryptophan oxygenase
Tryptophan pyrrolase
Tryptophanase
Protein vermilion
Tryptophan oxygenase
Tryptophan pyrrolase
Tryptophanase
Protein vermilion
Location in the cell
Cytoplasmic Reliability : 1.94 Nuclear Reliability : 1.622
Sequence
CDS
ATGGCGTGTCCTATGAGATCAGCGATCGACGAAATAAATGGCCAAGAAGGCGCGCACCTTGGTAACGAGGCCGGCATGCTATACGGGGAATATTTGATGCTGGATAAACTGCTGTCTGCGCAGAGGATGCTCAGCGCTGAGTCCTCTAAACCTGTACACGATGAGCATCTCTTCATCATCACACACCAAGCCTACGAACTCTGGTTCAAGCAGATAATATTTGAAGTGGATTCTGTGAGGGCATTACTTAACGTGGAAGGCTTGGACGAAAGCCACACCATGGAGATATTGAAGAGACTCAACAGGATTGTGCTCATCTTGAAACTGCTGGTGGATCAAGTGATGATTTTGGAGACGATGACACCGCTCGACTTCATGGATTTCAGGCACTATCTCCGCCCAGCATCAGGTTTCCAGAGCTTGCAGTTCAGACTACTAGAAAACAAGCTTGGTTTGAAACAAGCCCTGCGCGTGAAGTACAATCAAAATTATCAAACCGTGTTTGGCGACGACCCTGAAGCTATGGACTCCTTACAAAAATCTGAACAAGAACCTGCACTGCTAGCGCTGATCGAGCGTTGGCTGGAGCGTACTCCCGGATTGAACACGCACGGGTTCAACTTCTGGGGGAAATTCCAGGCAACAGTCAATAAAATGCTGAAGGAAGATATAGAGACTGCTAATCTTGAGTCCATAGACGCTGTGCGCCGCCATCGGCTTCAAGATGTGGAGAATAGAAGGGAGATTTATCGTTCGATATTTGATCCGACTGTGCATGACGCGCTCAGGTCCAGAGGAGAAAGGAGGCTGTCACATAGAGCACTCCAAGGTGCAATCATGATCACGTTTTACCGCGACGAGCCTCGGTTCTCGCAGCCGCATCAGCTCCTCACTCTGCTGATGGACATCGACAGTCTCATCACGAAATGGCGTTACAACCACGTTATAATGGTGCAAAGGATGATCGGGTCGCAACAGCTCGGGACCGGGGGCTCGTCTGGCTATCAGTATCTACGGTCAACATTAAGCGATCGTTACAAAGTCTTTCTTGATCTGTTCAATTTGTCCACGTTCCTGTTGCCTCGTGCCCTAATACCGCCACTGGACGAAGACATGAAACGCAGCCTCAATCTCACATGGGGAGACAACGTCAGGGAAAACGGAGATGAGACGACCCCGCAAAACGGCCTTGAAGCTTCTTTATAA
Protein
MACPMRSAIDEINGQEGAHLGNEAGMLYGEYLMLDKLLSAQRMLSAESSKPVHDEHLFIITHQAYELWFKQIIFEVDSVRALLNVEGLDESHTMEILKRLNRIVLILKLLVDQVMILETMTPLDFMDFRHYLRPASGFQSLQFRLLENKLGLKQALRVKYNQNYQTVFGDDPEAMDSLQKSEQEPALLALIERWLERTPGLNTHGFNFWGKFQATVNKMLKEDIETANLESIDAVRRHRLQDVENRREIYRSIFDPTVHDALRSRGERRLSHRALQGAIMITFYRDEPRFSQPHQLLTLLMDIDSLITKWRYNHVIMVQRMIGSQQLGTGGSSGYQYLRSTLSDRYKVFLDLFNLSTFLLPRALIPPLDEDMKRSLNLTWGDNVRENGDETTPQNGLEASL
Summary
Description
Heme-dependent dioxygenase that catalyzes the oxidative cleavage of the L-tryptophan (L-Trp) pyrrole ring and converts L-tryptophan to N-formyl-L-kynurenine. Catalyzes the oxidative cleavage of the indole moiety.
Heme-dependent dioxygenase that catalyzes the oxidative cleavage of the L-tryptophan (L-Trp) pyrrole ring and converts L-tryptophan to N-formyl-L-kynurenine. Catalyzes the oxidative cleavage of the indole moiety (By similarity). Required for normal eye pigmentation.
Heme-dependent dioxygenase that catalyzes the oxidative cleavage of the L-tryptophan (L-Trp) pyrrole ring and converts L-tryptophan to N-formyl-L-kynurenine. Catalyzes the oxidative cleavage of the indole moiety (PubMed:23333332). Required during larval growth to control the level of potentially harmful free tryptophan in the hemolymph. In the adult the same reaction is the first step in the ommochrome biosynthetic pathway (PubMed:2108317).
Heme-dependent dioxygenase that catalyzes the oxidative cleavage of the L-tryptophan (L-Trp) pyrrole ring and converts L-tryptophan to N-formyl-L-kynurenine. Catalyzes the oxidative cleavage of the indole moiety (By similarity). Required for normal eye pigmentation.
Heme-dependent dioxygenase that catalyzes the oxidative cleavage of the L-tryptophan (L-Trp) pyrrole ring and converts L-tryptophan to N-formyl-L-kynurenine. Catalyzes the oxidative cleavage of the indole moiety (PubMed:23333332). Required during larval growth to control the level of potentially harmful free tryptophan in the hemolymph. In the adult the same reaction is the first step in the ommochrome biosynthetic pathway (PubMed:2108317).
Catalytic Activity
L-tryptophan + O2 = N-formyl-L-kynurenine
Cofactor
heme
Biophysicochemical Properties
Optimum pH is 8.0.
Subunit
Homotetramer. Dimer of dimers.
Similarity
Belongs to the tryptophan 2,3-dioxygenase family.
Belongs to the peptidase C1 family.
Belongs to the peptidase C1 family.
Keywords
Complete proteome
Dioxygenase
Heme
Iron
Metal-binding
Oxidoreductase
Reference proteome
Tryptophan catabolism
3D-structure
Feature
chain Tryptophan 2,3-dioxygenase
Uniprot
H9JIC6
A0A2R4AL92
A0A194Q4A4
A0A2A4JTY2
S4PSA4
Q6T8S3
+ More
A0A212EZ73 A0A0N1INH1 A0A182QDX5 A0A1I8JW41 A0A182V5A1 A0A182K089 A0A182U7N7 A0A2M4BQI7 A0A182PUY7 A0A182VZT9 A0A2M4ASM5 A0A182FQY9 W5JG23 A0A182LE44 A0A182IIH8 O77457 A0A182Y3Z9 A0A2M4BQI3 A0A182MEF9 A0A2M4BQI6 A0A182NR48 A0A182R450 U5EUL1 A0A2M3ZJ82 A0A2M3ZJ61 A0A182H2Y2 A0A182ITI0 Q17P71 A0A1Q3G4E7 A0A2P8XV95 A0A1Y1L5F7 A0A0A1WSG4 A0A023ET96 A0A034VM38 A0A0K8UXS2 A0A0L0C3U8 A0A067RAS9 A0A336LP37 A0A1J1IDK9 Q95NN1 W8BZZ3 B4L629 D1ZZE3 A0A2J7QIW9 T1PGE3 D3TPI1 A0A1B0APM4 A0A1I8N0T4 A0A1A9YNV9 B3MQP7 O17441 A0A336LNM4 J3JYL8 B4MSH7 B4M818 B3NVC6 Q29I03 B4H4U3 P20351 B4JKK1 Q24630 B4IDV8 A0A1B0AEC0 A0A1A9VEK9 B4PYW0 A0A1W4UND5 A0A3B0KZT5 A0A1I8P805 A0A3B0KW42 A0A1B0FCL6 Q9NGA5 G3CRK2 A0A3B0KSD2 A0A146LPU6 A0A0A9XSB5 K7J8W4 A0A0M4ETM2 R4FKM1 A0A3S9Y2C2 A0A224XRU2 Q2LD53 A0A154NXN2 A0A0L7R814 A0A3B0KLW9 A0A023F8V1 A0A026W1I7 A0A088AV84 A0A2A3ES24 C8YQG0 E0VBN6 A0A0N7Z8R7 A0A1W4WUA4 A0A084VRJ5
A0A212EZ73 A0A0N1INH1 A0A182QDX5 A0A1I8JW41 A0A182V5A1 A0A182K089 A0A182U7N7 A0A2M4BQI7 A0A182PUY7 A0A182VZT9 A0A2M4ASM5 A0A182FQY9 W5JG23 A0A182LE44 A0A182IIH8 O77457 A0A182Y3Z9 A0A2M4BQI3 A0A182MEF9 A0A2M4BQI6 A0A182NR48 A0A182R450 U5EUL1 A0A2M3ZJ82 A0A2M3ZJ61 A0A182H2Y2 A0A182ITI0 Q17P71 A0A1Q3G4E7 A0A2P8XV95 A0A1Y1L5F7 A0A0A1WSG4 A0A023ET96 A0A034VM38 A0A0K8UXS2 A0A0L0C3U8 A0A067RAS9 A0A336LP37 A0A1J1IDK9 Q95NN1 W8BZZ3 B4L629 D1ZZE3 A0A2J7QIW9 T1PGE3 D3TPI1 A0A1B0APM4 A0A1I8N0T4 A0A1A9YNV9 B3MQP7 O17441 A0A336LNM4 J3JYL8 B4MSH7 B4M818 B3NVC6 Q29I03 B4H4U3 P20351 B4JKK1 Q24630 B4IDV8 A0A1B0AEC0 A0A1A9VEK9 B4PYW0 A0A1W4UND5 A0A3B0KZT5 A0A1I8P805 A0A3B0KW42 A0A1B0FCL6 Q9NGA5 G3CRK2 A0A3B0KSD2 A0A146LPU6 A0A0A9XSB5 K7J8W4 A0A0M4ETM2 R4FKM1 A0A3S9Y2C2 A0A224XRU2 Q2LD53 A0A154NXN2 A0A0L7R814 A0A3B0KLW9 A0A023F8V1 A0A026W1I7 A0A088AV84 A0A2A3ES24 C8YQG0 E0VBN6 A0A0N7Z8R7 A0A1W4WUA4 A0A084VRJ5
EC Number
1.13.11.11
Pubmed
19121390
26354079
23622113
15861231
22118469
20920257
+ More
23761445 20966253 8969464 12364791 25244985 26483478 17212352 17510324 29403074 28004739 25830018 24945155 25348373 26108605 24845553 11805058 24495485 17994087 18362917 19820115 20353571 25315136 9576938 22516182 23537049 15632085 2108317 10731132 12537572 12537569 23333332 7672574 10823947 26823975 25401762 20075255 30588650 25474469 24508170 30249741 19506309 20566863 27129103 24438588
23761445 20966253 8969464 12364791 25244985 26483478 17212352 17510324 29403074 28004739 25830018 24945155 25348373 26108605 24845553 11805058 24495485 17994087 18362917 19820115 20353571 25315136 9576938 22516182 23537049 15632085 2108317 10731132 12537572 12537569 23333332 7672574 10823947 26823975 25401762 20075255 30588650 25474469 24508170 30249741 19506309 20566863 27129103 24438588
EMBL
BABH01022310
MF598173
MG976796
AVR75813.1
AVR75814.1
KQ459585
+ More
KPI98240.1 NWSH01000678 PCG74852.1 GAIX01014028 JAA78532.1 AY427951 AAR24625.1 AGBW02011349 OWR46799.1 KQ461193 KPJ06985.1 AXCN02001476 GGFJ01006206 MBW55347.1 GGFK01010458 MBW43779.1 ADMH02001265 ETN63322.1 APCN01000245 L76432 L76433 AAAB01008859 GGFJ01006204 MBW55345.1 AXCM01000144 GGFJ01006205 MBW55346.1 GANO01002268 JAB57603.1 GGFM01007822 MBW28573.1 GGFM01007825 MBW28576.1 JXUM01024210 JXUM01024211 KQ560683 KXJ81288.1 AF325458 CH477193 GFDL01000381 JAV34664.1 PYGN01001295 PSN35923.1 GEZM01066971 JAV67610.1 GBXI01012273 JAD02019.1 GAPW01001994 JAC11604.1 GAKP01014561 JAC44391.1 GDHF01020842 JAI31472.1 JRES01000940 KNC27013.1 KK852773 KDR16811.1 UFQS01000095 UFQT01000095 SSW99300.1 SSX19680.1 CVRI01000047 CRK97636.1 AY052390 AY052392 GAMC01004257 JAC02299.1 CH933812 KQ971338 EFA02356.1 NEVH01013570 PNF28534.1 KA647210 AFP61839.1 EZ423333 ADD19609.1 JXJN01001487 AF028834 CH902621 AF028835 AAC24240.1 SSW99302.1 SSX19682.1 BT128347 KB632166 AEE63305.1 ERL89336.1 CH963851 CH940653 CH954180 CH379064 CH479209 M34147 AE014298 AY051478 CH916370 U27204 CM000366 CH480830 CM000162 OUUW01000025 SPP89618.1 SPP89621.1 CCAG010004530 AF255325 AAF68620.1 HQ157970 ADN78296.1 SPP89619.1 GDHC01009857 JAQ08772.1 GBHO01020750 JAG22854.1 AAZX01019176 CP012528 ALC49902.1 ACPB03015151 GAHY01001788 JAA75722.1 MH806846 AZS64103.1 GFTR01005727 JAW10699.1 DQ335251 KQ434778 KZC04435.1 KQ414636 KOC67020.1 SPP89620.1 GBBI01000870 JAC17842.1 KK107488 QOIP01000012 EZA49883.1 RLU15875.1 KZ288192 PBC34314.1 FJ696203 FJ696204 FJ696205 FJ696206 ACV81725.1 DS235033 EEB10792.1 GDKW01002595 JAI54000.1 ATLV01015693 ATLV01015694 ATLV01015695 ATLV01015696 KE525027 KFB40589.1
KPI98240.1 NWSH01000678 PCG74852.1 GAIX01014028 JAA78532.1 AY427951 AAR24625.1 AGBW02011349 OWR46799.1 KQ461193 KPJ06985.1 AXCN02001476 GGFJ01006206 MBW55347.1 GGFK01010458 MBW43779.1 ADMH02001265 ETN63322.1 APCN01000245 L76432 L76433 AAAB01008859 GGFJ01006204 MBW55345.1 AXCM01000144 GGFJ01006205 MBW55346.1 GANO01002268 JAB57603.1 GGFM01007822 MBW28573.1 GGFM01007825 MBW28576.1 JXUM01024210 JXUM01024211 KQ560683 KXJ81288.1 AF325458 CH477193 GFDL01000381 JAV34664.1 PYGN01001295 PSN35923.1 GEZM01066971 JAV67610.1 GBXI01012273 JAD02019.1 GAPW01001994 JAC11604.1 GAKP01014561 JAC44391.1 GDHF01020842 JAI31472.1 JRES01000940 KNC27013.1 KK852773 KDR16811.1 UFQS01000095 UFQT01000095 SSW99300.1 SSX19680.1 CVRI01000047 CRK97636.1 AY052390 AY052392 GAMC01004257 JAC02299.1 CH933812 KQ971338 EFA02356.1 NEVH01013570 PNF28534.1 KA647210 AFP61839.1 EZ423333 ADD19609.1 JXJN01001487 AF028834 CH902621 AF028835 AAC24240.1 SSW99302.1 SSX19682.1 BT128347 KB632166 AEE63305.1 ERL89336.1 CH963851 CH940653 CH954180 CH379064 CH479209 M34147 AE014298 AY051478 CH916370 U27204 CM000366 CH480830 CM000162 OUUW01000025 SPP89618.1 SPP89621.1 CCAG010004530 AF255325 AAF68620.1 HQ157970 ADN78296.1 SPP89619.1 GDHC01009857 JAQ08772.1 GBHO01020750 JAG22854.1 AAZX01019176 CP012528 ALC49902.1 ACPB03015151 GAHY01001788 JAA75722.1 MH806846 AZS64103.1 GFTR01005727 JAW10699.1 DQ335251 KQ434778 KZC04435.1 KQ414636 KOC67020.1 SPP89620.1 GBBI01000870 JAC17842.1 KK107488 QOIP01000012 EZA49883.1 RLU15875.1 KZ288192 PBC34314.1 FJ696203 FJ696204 FJ696205 FJ696206 ACV81725.1 DS235033 EEB10792.1 GDKW01002595 JAI54000.1 ATLV01015693 ATLV01015694 ATLV01015695 ATLV01015696 KE525027 KFB40589.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000075886
+ More
UP000076407 UP000075903 UP000075881 UP000075902 UP000075885 UP000075920 UP000069272 UP000000673 UP000075882 UP000075840 UP000007062 UP000076408 UP000075883 UP000075884 UP000075900 UP000069940 UP000249989 UP000075880 UP000008820 UP000245037 UP000037069 UP000027135 UP000183832 UP000009192 UP000007266 UP000235965 UP000092460 UP000095301 UP000092443 UP000007801 UP000030742 UP000007798 UP000008792 UP000008711 UP000001819 UP000008744 UP000000803 UP000001070 UP000000304 UP000001292 UP000092445 UP000078200 UP000002282 UP000192221 UP000268350 UP000095300 UP000092444 UP000002358 UP000092553 UP000015103 UP000076502 UP000053825 UP000053097 UP000279307 UP000005203 UP000242457 UP000009046 UP000192223 UP000030765
UP000076407 UP000075903 UP000075881 UP000075902 UP000075885 UP000075920 UP000069272 UP000000673 UP000075882 UP000075840 UP000007062 UP000076408 UP000075883 UP000075884 UP000075900 UP000069940 UP000249989 UP000075880 UP000008820 UP000245037 UP000037069 UP000027135 UP000183832 UP000009192 UP000007266 UP000235965 UP000092460 UP000095301 UP000092443 UP000007801 UP000030742 UP000007798 UP000008792 UP000008711 UP000001819 UP000008744 UP000000803 UP000001070 UP000000304 UP000001292 UP000092445 UP000078200 UP000002282 UP000192221 UP000268350 UP000095300 UP000092444 UP000002358 UP000092553 UP000015103 UP000076502 UP000053825 UP000053097 UP000279307 UP000005203 UP000242457 UP000009046 UP000192223 UP000030765
PRIDE
Interpro
IPR037217
Trp/Indoleamine_2_3_dOase-like
+ More
IPR004981 Trp_2_3_dOase
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR038765 Papain-like_cys_pep_sf
IPR039417 Peptidase_C1A_papain-like
IPR025660 Pept_his_AS
IPR013201 Prot_inhib_I29
IPR000668 Peptidase_C1A_C
IPR000169 Pept_cys_AS
IPR002942 S4_RNA-bd
IPR018079 Ribosomal_S4_CS
IPR036986 S4_RNA-bd_sf
IPR005710 Ribosomal_S4/S9_euk/arc
IPR001912 Ribosomal_S4/S9_N
IPR004981 Trp_2_3_dOase
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR038765 Papain-like_cys_pep_sf
IPR039417 Peptidase_C1A_papain-like
IPR025660 Pept_his_AS
IPR013201 Prot_inhib_I29
IPR000668 Peptidase_C1A_C
IPR000169 Pept_cys_AS
IPR002942 S4_RNA-bd
IPR018079 Ribosomal_S4_CS
IPR036986 S4_RNA-bd_sf
IPR005710 Ribosomal_S4/S9_euk/arc
IPR001912 Ribosomal_S4/S9_N
Gene 3D
ProteinModelPortal
H9JIC6
A0A2R4AL92
A0A194Q4A4
A0A2A4JTY2
S4PSA4
Q6T8S3
+ More
A0A212EZ73 A0A0N1INH1 A0A182QDX5 A0A1I8JW41 A0A182V5A1 A0A182K089 A0A182U7N7 A0A2M4BQI7 A0A182PUY7 A0A182VZT9 A0A2M4ASM5 A0A182FQY9 W5JG23 A0A182LE44 A0A182IIH8 O77457 A0A182Y3Z9 A0A2M4BQI3 A0A182MEF9 A0A2M4BQI6 A0A182NR48 A0A182R450 U5EUL1 A0A2M3ZJ82 A0A2M3ZJ61 A0A182H2Y2 A0A182ITI0 Q17P71 A0A1Q3G4E7 A0A2P8XV95 A0A1Y1L5F7 A0A0A1WSG4 A0A023ET96 A0A034VM38 A0A0K8UXS2 A0A0L0C3U8 A0A067RAS9 A0A336LP37 A0A1J1IDK9 Q95NN1 W8BZZ3 B4L629 D1ZZE3 A0A2J7QIW9 T1PGE3 D3TPI1 A0A1B0APM4 A0A1I8N0T4 A0A1A9YNV9 B3MQP7 O17441 A0A336LNM4 J3JYL8 B4MSH7 B4M818 B3NVC6 Q29I03 B4H4U3 P20351 B4JKK1 Q24630 B4IDV8 A0A1B0AEC0 A0A1A9VEK9 B4PYW0 A0A1W4UND5 A0A3B0KZT5 A0A1I8P805 A0A3B0KW42 A0A1B0FCL6 Q9NGA5 G3CRK2 A0A3B0KSD2 A0A146LPU6 A0A0A9XSB5 K7J8W4 A0A0M4ETM2 R4FKM1 A0A3S9Y2C2 A0A224XRU2 Q2LD53 A0A154NXN2 A0A0L7R814 A0A3B0KLW9 A0A023F8V1 A0A026W1I7 A0A088AV84 A0A2A3ES24 C8YQG0 E0VBN6 A0A0N7Z8R7 A0A1W4WUA4 A0A084VRJ5
A0A212EZ73 A0A0N1INH1 A0A182QDX5 A0A1I8JW41 A0A182V5A1 A0A182K089 A0A182U7N7 A0A2M4BQI7 A0A182PUY7 A0A182VZT9 A0A2M4ASM5 A0A182FQY9 W5JG23 A0A182LE44 A0A182IIH8 O77457 A0A182Y3Z9 A0A2M4BQI3 A0A182MEF9 A0A2M4BQI6 A0A182NR48 A0A182R450 U5EUL1 A0A2M3ZJ82 A0A2M3ZJ61 A0A182H2Y2 A0A182ITI0 Q17P71 A0A1Q3G4E7 A0A2P8XV95 A0A1Y1L5F7 A0A0A1WSG4 A0A023ET96 A0A034VM38 A0A0K8UXS2 A0A0L0C3U8 A0A067RAS9 A0A336LP37 A0A1J1IDK9 Q95NN1 W8BZZ3 B4L629 D1ZZE3 A0A2J7QIW9 T1PGE3 D3TPI1 A0A1B0APM4 A0A1I8N0T4 A0A1A9YNV9 B3MQP7 O17441 A0A336LNM4 J3JYL8 B4MSH7 B4M818 B3NVC6 Q29I03 B4H4U3 P20351 B4JKK1 Q24630 B4IDV8 A0A1B0AEC0 A0A1A9VEK9 B4PYW0 A0A1W4UND5 A0A3B0KZT5 A0A1I8P805 A0A3B0KW42 A0A1B0FCL6 Q9NGA5 G3CRK2 A0A3B0KSD2 A0A146LPU6 A0A0A9XSB5 K7J8W4 A0A0M4ETM2 R4FKM1 A0A3S9Y2C2 A0A224XRU2 Q2LD53 A0A154NXN2 A0A0L7R814 A0A3B0KLW9 A0A023F8V1 A0A026W1I7 A0A088AV84 A0A2A3ES24 C8YQG0 E0VBN6 A0A0N7Z8R7 A0A1W4WUA4 A0A084VRJ5
PDB
4HKA
E-value=4.67809e-138,
Score=1259
Ontologies
KEGG
PATHWAY
GO
PANTHER
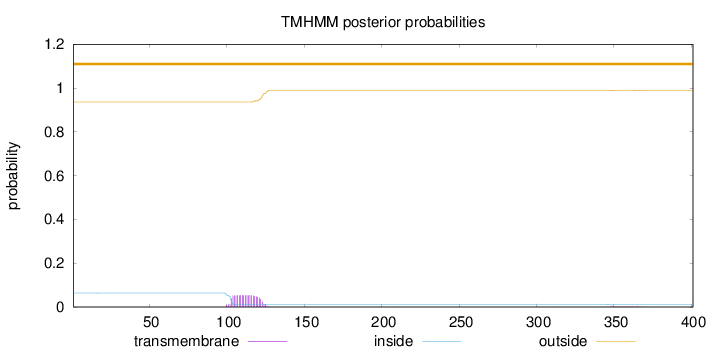
Topology
Length:
401
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.13216
Exp number, first 60 AAs:
0.00176
Total prob of N-in:
0.06366
outside
1 - 401
Population Genetic Test Statistics
Pi
181.352233
Theta
162.253402
Tajima's D
0.234518
CLR
0.62677
CSRT
0.438778061096945
Interpretation
Uncertain
