Gene
KWMTBOMO08349
Pre Gene Modal
BGIBMGA009273
Annotation
PREDICTED:_transient-receptor-potential-like_protein_[Amyelois_transitella]
Full name
Transient-receptor-potential-like protein
Location in the cell
Cytoplasmic Reliability : 2.652
Sequence
CDS
ATGGCCGAGAAGAAGGATTTGGAAGCCGGCGACCCTGAATTGGAATGCGTCCGGAAACCAATGCCGTTACCGGTGCTCCCAAAACCACTTACTTTGGAAGAAAAGAAGTATTTGCTGGCAGTCGAGAGAGGAGATATGGCAAATGTGAGAAGATTACTCCAAAAAGGCCACCGCAAAAAACACATCGACATCAATTGCGTAGACTCTCTCGGTCGCGGGGCTTTGACCTTGTCAATTGACGGGGAGAACTTAGAGATGGTGGAACTCCTCGTCATCATGGGAGTGGAAACAAGAGACGCTTTGCTGCAAGCAATCAACGCGGAGTTTGTTGAAGCAGTTGAATTGTTACTAGAGCATGAAGAGTTAATTCACAAAGACGGTGAACCATACAGCTGGCAAAAAGTTGATCCAAACACAGCAATGTTCACTCCGGACATAACACCTCTGATGCTGGCGGCTCATAAGAACAACTATGAAATAATCAAGATACTTCTAGATCGTGGTGCCACTTTACCTAACCCTCATGACGTCAGATGTGGCTGCGAGGACTGTATCCGACAATCTACTGAGGACTCTCTGCGACACTCACTGGCACGCCTCAATGAATATCGAGCTTTGGCATCTCCGTCGCTCATAGCTCTGTCTTCTACAGATCCTATATTAACAGCCTTCGAATTGTCTTGGGAACTGAGGAATTTAGCCTTTGCTGAACAGGAAAGTAAAGCGGAATACCTGGAACTACGGAGACAGGCGCAACTGTTTGCTGTAGATCTGCTCGATCAATCCCGAAGCTCCCAAGAGCTGGCCATCATTCTCAATCATGATCCTGACGAACCAGCCTTTGTTGACGGAGAACACATGAAATTAGCACGTTTGGAATTGGCCATTGATTTTAAGCAGAAAAAGTTCGTAGCTCACCCTAACATCCAACAATTGCTGGCCTCTATCTGGTACGAAGGTGTCCCTGGATTCCGTCGTAAAACTACAATGGAGAAGATCATGATCATCTGTCGCGTCGCCCTATTGTTCCCTTTCTACTGCACTCTGTACATGATAGCCCCAAACTGCGCCACCGGAAAACTAATGAGGAAACCTTTCATGAAGTTCTTAATTCACGCTTCTAGTTACTTATTCTTTTTGTGTAAGTAA
Protein
MAEKKDLEAGDPELECVRKPMPLPVLPKPLTLEEKKYLLAVERGDMANVRRLLQKGHRKKHIDINCVDSLGRGALTLSIDGENLEMVELLVIMGVETRDALLQAINAEFVEAVELLLEHEELIHKDGEPYSWQKVDPNTAMFTPDITPLMLAAHKNNYEIIKILLDRGATLPNPHDVRCGCEDCIRQSTEDSLRHSLARLNEYRALASPSLIALSSTDPILTAFELSWELRNLAFAEQESKAEYLELRRQAQLFAVDLLDQSRSSQELAIILNHDPDEPAFVDGEHMKLARLELAIDFKQKKFVAHPNIQQLLASIWYEGVPGFRRKTTMEKIMIICRVALLFPFYCTLYMIAPNCATGKLMRKPFMKFLIHASSYLFFLCK
Summary
Description
A light-sensitive calcium channel that is required for inositide-mediated Ca(2+) entry in the retina during phospholipase C (PLC)-mediated phototransduction. Required for vision in the dark and in dim light. Binds calmodulin. Trp and trpl act together in the light response, although it is unclear whether as heteromultimers or distinct units. Also forms a functional cation channel with Trpgamma. Activated by fatty acids, metabolic stress, inositols and GTP-binding proteins.
Subunit
Forms heteromultimers with Trpgamma and, to a lower extent, with trp. Interacts with FKBP59 in vivo and is found in the inaD signaling complex.
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Belongs to the transient receptor (TC 1.A.4) family. STrpC subfamily.
Belongs to the transient receptor (TC 1.A.4) family. STrpC subfamily.
Keywords
ANK repeat
Calcium
Calcium channel
Calcium transport
Calmodulin-binding
Complete proteome
Ion channel
Ion transport
Membrane
Reference proteome
Repeat
Sensory transduction
Transmembrane
Transmembrane helix
Transport
Vision
Feature
chain Transient-receptor-potential-like protein
Uniprot
H9JIC3
A0A2H1VKM1
A0A3S2NXY7
A0A2A4J1F1
A0A194PYX6
A0A0N1ICM2
+ More
A0A212EM72 A0A182T045 Q179M5 A0A1Q3FS27 A0A1S4FB01 B0WEN0 A0A2M4BBT8 A0A2M4BCG6 Q7PTP0 A0A182JLW8 M4N9U7 D2A1G1 A0A084WLR8 W5J1J3 A0A182FTJ6 A0A182TQV7 A0A182VI22 A0A453Z0C9 A0A182LXL9 A0A182KUV4 A0A182X327 A0A182I0Y2 A0A182PBX0 A0A182QP93 A0A182YA85 A0A182RLY7 T1DPY1 A0A2M4A8K7 A0A2M4A834 A0A2M4A8I3 A0A2M4A843 T1H886 A0A182K248 A0A182WEB6 A0A182NPI7 A0A336L2Z3 A0A182G1G9 A0A182HA26 A0A384T274 A0A0K8SKT6 A0A1B6LZM0 A0A1J1IYR4 A0A0A9WXN9 A0A1B6EER2 A0A0L0C5K2 D3TKW3 A0A1A9X4C4 A0A0A1WZ79 A0A0K8WLZ2 A0A1W4UZP9 A0A067RIF5 T1PFK2 A0A034W307 A0A1I8N2Y0 W8ABV7 W8B6G5 A0A146LMA2 B3MCR1 A0A1I8P5H8 A0A1B0A2P7 P48994 B4HT37 A0A1B0BHH4 B3N6W7 A0A1A9V4N0 B4NWK1 A0A0R1DTT2 B4GGD2 A0A0R3NUP5 Q28Y01 A0A0R3NP54 A0A0R3NPZ4 B4MS74 A0A3B0J0C2 A0A0J9TXH9 A0A0J9R904 B4JVY3 B4LJB1 U4UIT9 A0A0Q9XAA9 B4KTU2 A0A1B0GFH5 A0A2H4WAN6 N6TP96 A0A0J9R907 W5JJ05 A0A0R3NVH0 A0A0P5U0B8 X1X233 A0A182HBB8 A0A0P5WTL1 A0A182MJ53 A0A0P5NIX2
A0A212EM72 A0A182T045 Q179M5 A0A1Q3FS27 A0A1S4FB01 B0WEN0 A0A2M4BBT8 A0A2M4BCG6 Q7PTP0 A0A182JLW8 M4N9U7 D2A1G1 A0A084WLR8 W5J1J3 A0A182FTJ6 A0A182TQV7 A0A182VI22 A0A453Z0C9 A0A182LXL9 A0A182KUV4 A0A182X327 A0A182I0Y2 A0A182PBX0 A0A182QP93 A0A182YA85 A0A182RLY7 T1DPY1 A0A2M4A8K7 A0A2M4A834 A0A2M4A8I3 A0A2M4A843 T1H886 A0A182K248 A0A182WEB6 A0A182NPI7 A0A336L2Z3 A0A182G1G9 A0A182HA26 A0A384T274 A0A0K8SKT6 A0A1B6LZM0 A0A1J1IYR4 A0A0A9WXN9 A0A1B6EER2 A0A0L0C5K2 D3TKW3 A0A1A9X4C4 A0A0A1WZ79 A0A0K8WLZ2 A0A1W4UZP9 A0A067RIF5 T1PFK2 A0A034W307 A0A1I8N2Y0 W8ABV7 W8B6G5 A0A146LMA2 B3MCR1 A0A1I8P5H8 A0A1B0A2P7 P48994 B4HT37 A0A1B0BHH4 B3N6W7 A0A1A9V4N0 B4NWK1 A0A0R1DTT2 B4GGD2 A0A0R3NUP5 Q28Y01 A0A0R3NP54 A0A0R3NPZ4 B4MS74 A0A3B0J0C2 A0A0J9TXH9 A0A0J9R904 B4JVY3 B4LJB1 U4UIT9 A0A0Q9XAA9 B4KTU2 A0A1B0GFH5 A0A2H4WAN6 N6TP96 A0A0J9R907 W5JJ05 A0A0R3NVH0 A0A0P5U0B8 X1X233 A0A182HBB8 A0A0P5WTL1 A0A182MJ53 A0A0P5NIX2
Pubmed
19121390
26354079
22118469
17510324
12364791
18362917
+ More
19820115 24438588 20920257 23761445 20966253 25244985 26483478 25401762 26108605 20353571 25830018 24845553 25348373 25315136 24495485 26823975 17994087 18057021 1314616 10731132 12537572 12537569 8670063 9215637 9494079 10371201 9930700 10908615 8918461 10896160 11514552 11931743 11707492 17550304 15632085 23185243 22936249 23537049 29182505
19820115 24438588 20920257 23761445 20966253 25244985 26483478 25401762 26108605 20353571 25830018 24845553 25348373 25315136 24495485 26823975 17994087 18057021 1314616 10731132 12537572 12537569 8670063 9215637 9494079 10371201 9930700 10908615 8918461 10896160 11514552 11931743 11707492 17550304 15632085 23185243 22936249 23537049 29182505
EMBL
BABH01022308
ODYU01003065
SOQ41367.1
RSAL01000028
RVE51848.1
NWSH01004108
+ More
PCG65488.1 KQ459585 KPI98238.1 KQ459786 KPJ19920.1 AGBW02013881 OWR42592.1 CH477348 EAT42934.1 GFDL01004651 JAV30394.1 DS231910 EDS45746.1 GGFJ01001365 MBW50506.1 GGFJ01001367 MBW50508.1 AAAB01008796 EAA03611.5 KC292630 AGG86915.1 KQ971338 EFA02859.1 ATLV01024272 KE525351 KFB51162.1 ADMH02002175 ETN58052.1 AXCM01007166 APCN01006537 AXCN02001829 GAMD01002929 JAA98661.1 GGFK01003627 MBW36948.1 GGFK01003642 MBW36963.1 GGFK01003597 MBW36918.1 GGFK01003628 MBW36949.1 ACPB03011940 ACPB03011941 UFQS01001431 UFQT01001431 SSX10849.1 SSX30529.1 JXUM01136915 JXUM01136916 JXUM01136917 KQ568500 KXJ68981.1 JXUM01030796 KQ560896 KXJ80443.1 KX249689 AOR81466.1 GBRD01011882 JAG53942.1 GEBQ01010897 JAT29080.1 CVRI01000063 CRL04302.1 GBHO01030377 JAG13227.1 GEDC01000871 JAS36427.1 JRES01000890 KNC27516.1 EZ422050 ADD18473.1 GBXI01010130 GBXI01008708 GBXI01004578 JAD04162.1 JAD05584.1 JAD09714.1 GDHF01005173 GDHF01001552 GDHF01000136 JAI47141.1 JAI50762.1 JAI52178.1 KK852485 KDR22808.1 KA646945 AFP61574.1 GAKP01010448 GAKP01010447 JAC48505.1 GAMC01021170 JAB85385.1 GAMC01021171 JAB85384.1 GDHC01010100 JAQ08529.1 CH902619 EDV37313.1 KPU76709.1 KPU76710.1 M88185 AE013599 BT001397 BT099607 CH480816 EDW47147.1 JXJN01014362 JXJN01014363 CH954177 EDV58216.1 CM000157 EDW89546.1 KRJ98521.1 KRJ98522.1 CH479183 EDW35552.1 CM000071 KRT02893.1 EAL26165.2 KRT02894.1 KRT02892.1 KRT02895.1 CH963850 EDW74963.1 OUUW01000001 SPP74037.1 CM002911 KMY92564.1 KMY92565.1 KMY92566.1 KMY92562.1 CH916375 EDV98121.1 CH940648 EDW60491.1 KRF79437.1 KRF79438.1 KRF79439.1 KRF79440.1 KB632375 ERL93959.1 CH933808 KRG05424.1 EDW10668.1 CCAG010006888 MG020535 AUC64091.1 APGK01013995 APGK01013996 APGK01013997 APGK01013998 APGK01013999 APGK01014000 KB739168 ENN82359.1 KMY92563.1 ADMH02001356 ETN62865.1 KRT02896.1 GDIP01119518 JAL84196.1 ABLF02021769 ABLF02021770 ABLF02021771 ABLF02052683 JXUM01004958 JXUM01004959 JXUM01004960 JXUM01004961 JXUM01004962 KQ560184 KXJ83949.1 GDIP01094025 JAM09690.1 AXCM01012156 GDIQ01144554 JAL07172.1
PCG65488.1 KQ459585 KPI98238.1 KQ459786 KPJ19920.1 AGBW02013881 OWR42592.1 CH477348 EAT42934.1 GFDL01004651 JAV30394.1 DS231910 EDS45746.1 GGFJ01001365 MBW50506.1 GGFJ01001367 MBW50508.1 AAAB01008796 EAA03611.5 KC292630 AGG86915.1 KQ971338 EFA02859.1 ATLV01024272 KE525351 KFB51162.1 ADMH02002175 ETN58052.1 AXCM01007166 APCN01006537 AXCN02001829 GAMD01002929 JAA98661.1 GGFK01003627 MBW36948.1 GGFK01003642 MBW36963.1 GGFK01003597 MBW36918.1 GGFK01003628 MBW36949.1 ACPB03011940 ACPB03011941 UFQS01001431 UFQT01001431 SSX10849.1 SSX30529.1 JXUM01136915 JXUM01136916 JXUM01136917 KQ568500 KXJ68981.1 JXUM01030796 KQ560896 KXJ80443.1 KX249689 AOR81466.1 GBRD01011882 JAG53942.1 GEBQ01010897 JAT29080.1 CVRI01000063 CRL04302.1 GBHO01030377 JAG13227.1 GEDC01000871 JAS36427.1 JRES01000890 KNC27516.1 EZ422050 ADD18473.1 GBXI01010130 GBXI01008708 GBXI01004578 JAD04162.1 JAD05584.1 JAD09714.1 GDHF01005173 GDHF01001552 GDHF01000136 JAI47141.1 JAI50762.1 JAI52178.1 KK852485 KDR22808.1 KA646945 AFP61574.1 GAKP01010448 GAKP01010447 JAC48505.1 GAMC01021170 JAB85385.1 GAMC01021171 JAB85384.1 GDHC01010100 JAQ08529.1 CH902619 EDV37313.1 KPU76709.1 KPU76710.1 M88185 AE013599 BT001397 BT099607 CH480816 EDW47147.1 JXJN01014362 JXJN01014363 CH954177 EDV58216.1 CM000157 EDW89546.1 KRJ98521.1 KRJ98522.1 CH479183 EDW35552.1 CM000071 KRT02893.1 EAL26165.2 KRT02894.1 KRT02892.1 KRT02895.1 CH963850 EDW74963.1 OUUW01000001 SPP74037.1 CM002911 KMY92564.1 KMY92565.1 KMY92566.1 KMY92562.1 CH916375 EDV98121.1 CH940648 EDW60491.1 KRF79437.1 KRF79438.1 KRF79439.1 KRF79440.1 KB632375 ERL93959.1 CH933808 KRG05424.1 EDW10668.1 CCAG010006888 MG020535 AUC64091.1 APGK01013995 APGK01013996 APGK01013997 APGK01013998 APGK01013999 APGK01014000 KB739168 ENN82359.1 KMY92563.1 ADMH02001356 ETN62865.1 KRT02896.1 GDIP01119518 JAL84196.1 ABLF02021769 ABLF02021770 ABLF02021771 ABLF02052683 JXUM01004958 JXUM01004959 JXUM01004960 JXUM01004961 JXUM01004962 KQ560184 KXJ83949.1 GDIP01094025 JAM09690.1 AXCM01012156 GDIQ01144554 JAL07172.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000075901 UP000008820 UP000002320 UP000007062 UP000075880 UP000007266 UP000030765 UP000000673 UP000069272 UP000075902 UP000075903 UP000075883 UP000075882 UP000076407 UP000075840 UP000075885 UP000075886 UP000076408 UP000075900 UP000015103 UP000075881 UP000075920 UP000075884 UP000069940 UP000249989 UP000183832 UP000037069 UP000091820 UP000192221 UP000027135 UP000095301 UP000007801 UP000095300 UP000092445 UP000000803 UP000001292 UP000092460 UP000008711 UP000078200 UP000002282 UP000008744 UP000001819 UP000007798 UP000268350 UP000001070 UP000008792 UP000030742 UP000009192 UP000092444 UP000019118 UP000007819
UP000075901 UP000008820 UP000002320 UP000007062 UP000075880 UP000007266 UP000030765 UP000000673 UP000069272 UP000075902 UP000075903 UP000075883 UP000075882 UP000076407 UP000075840 UP000075885 UP000075886 UP000076408 UP000075900 UP000015103 UP000075881 UP000075920 UP000075884 UP000069940 UP000249989 UP000183832 UP000037069 UP000091820 UP000192221 UP000027135 UP000095301 UP000007801 UP000095300 UP000092445 UP000000803 UP000001292 UP000092460 UP000008711 UP000078200 UP000002282 UP000008744 UP000001819 UP000007798 UP000268350 UP000001070 UP000008792 UP000030742 UP000009192 UP000092444 UP000019118 UP000007819
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
H9JIC3
A0A2H1VKM1
A0A3S2NXY7
A0A2A4J1F1
A0A194PYX6
A0A0N1ICM2
+ More
A0A212EM72 A0A182T045 Q179M5 A0A1Q3FS27 A0A1S4FB01 B0WEN0 A0A2M4BBT8 A0A2M4BCG6 Q7PTP0 A0A182JLW8 M4N9U7 D2A1G1 A0A084WLR8 W5J1J3 A0A182FTJ6 A0A182TQV7 A0A182VI22 A0A453Z0C9 A0A182LXL9 A0A182KUV4 A0A182X327 A0A182I0Y2 A0A182PBX0 A0A182QP93 A0A182YA85 A0A182RLY7 T1DPY1 A0A2M4A8K7 A0A2M4A834 A0A2M4A8I3 A0A2M4A843 T1H886 A0A182K248 A0A182WEB6 A0A182NPI7 A0A336L2Z3 A0A182G1G9 A0A182HA26 A0A384T274 A0A0K8SKT6 A0A1B6LZM0 A0A1J1IYR4 A0A0A9WXN9 A0A1B6EER2 A0A0L0C5K2 D3TKW3 A0A1A9X4C4 A0A0A1WZ79 A0A0K8WLZ2 A0A1W4UZP9 A0A067RIF5 T1PFK2 A0A034W307 A0A1I8N2Y0 W8ABV7 W8B6G5 A0A146LMA2 B3MCR1 A0A1I8P5H8 A0A1B0A2P7 P48994 B4HT37 A0A1B0BHH4 B3N6W7 A0A1A9V4N0 B4NWK1 A0A0R1DTT2 B4GGD2 A0A0R3NUP5 Q28Y01 A0A0R3NP54 A0A0R3NPZ4 B4MS74 A0A3B0J0C2 A0A0J9TXH9 A0A0J9R904 B4JVY3 B4LJB1 U4UIT9 A0A0Q9XAA9 B4KTU2 A0A1B0GFH5 A0A2H4WAN6 N6TP96 A0A0J9R907 W5JJ05 A0A0R3NVH0 A0A0P5U0B8 X1X233 A0A182HBB8 A0A0P5WTL1 A0A182MJ53 A0A0P5NIX2
A0A212EM72 A0A182T045 Q179M5 A0A1Q3FS27 A0A1S4FB01 B0WEN0 A0A2M4BBT8 A0A2M4BCG6 Q7PTP0 A0A182JLW8 M4N9U7 D2A1G1 A0A084WLR8 W5J1J3 A0A182FTJ6 A0A182TQV7 A0A182VI22 A0A453Z0C9 A0A182LXL9 A0A182KUV4 A0A182X327 A0A182I0Y2 A0A182PBX0 A0A182QP93 A0A182YA85 A0A182RLY7 T1DPY1 A0A2M4A8K7 A0A2M4A834 A0A2M4A8I3 A0A2M4A843 T1H886 A0A182K248 A0A182WEB6 A0A182NPI7 A0A336L2Z3 A0A182G1G9 A0A182HA26 A0A384T274 A0A0K8SKT6 A0A1B6LZM0 A0A1J1IYR4 A0A0A9WXN9 A0A1B6EER2 A0A0L0C5K2 D3TKW3 A0A1A9X4C4 A0A0A1WZ79 A0A0K8WLZ2 A0A1W4UZP9 A0A067RIF5 T1PFK2 A0A034W307 A0A1I8N2Y0 W8ABV7 W8B6G5 A0A146LMA2 B3MCR1 A0A1I8P5H8 A0A1B0A2P7 P48994 B4HT37 A0A1B0BHH4 B3N6W7 A0A1A9V4N0 B4NWK1 A0A0R1DTT2 B4GGD2 A0A0R3NUP5 Q28Y01 A0A0R3NP54 A0A0R3NPZ4 B4MS74 A0A3B0J0C2 A0A0J9TXH9 A0A0J9R904 B4JVY3 B4LJB1 U4UIT9 A0A0Q9XAA9 B4KTU2 A0A1B0GFH5 A0A2H4WAN6 N6TP96 A0A0J9R907 W5JJ05 A0A0R3NVH0 A0A0P5U0B8 X1X233 A0A182HBB8 A0A0P5WTL1 A0A182MJ53 A0A0P5NIX2
PDB
6G1K
E-value=3.85762e-81,
Score=767
Ontologies
GO
GO:0005262
GO:0016021
GO:0006811
GO:0034703
GO:0006828
GO:0070679
GO:0005887
GO:0015279
GO:0051480
GO:0070588
GO:0016020
GO:0046982
GO:0030425
GO:0010461
GO:0005516
GO:0016027
GO:0007589
GO:0007605
GO:0042802
GO:0019722
GO:0035997
GO:0071454
GO:0007603
GO:0006812
GO:0050908
GO:0005886
GO:0006816
GO:0015075
GO:0005261
GO:0009416
GO:0016028
GO:0005515
GO:0005216
GO:0005839
GO:0051603
GO:0030145
GO:0006412
GO:0016876
GO:0043039
PANTHER
Topology
Subcellular location
Membrane
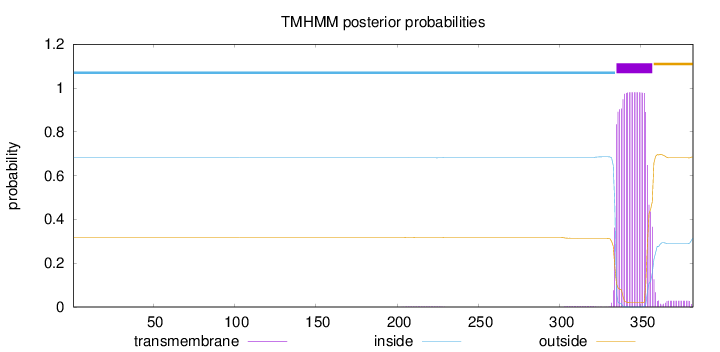
Length:
382
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.32642
Exp number, first 60 AAs:
0
Total prob of N-in:
0.68118
inside
1 - 334
TMhelix
335 - 357
outside
358 - 382
Population Genetic Test Statistics
Pi
17.466273
Theta
160.515432
Tajima's D
0.201529
CLR
0.647108
CSRT
0.431878406079696
Interpretation
Uncertain
