Gene
KWMTBOMO08347
Pre Gene Modal
BGIBMGA009272
Annotation
PREDICTED:_transient_receptor_potential_protein_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.957
Sequence
CDS
ATGTGGTATTATGCTGAACTAGAAAAAGACAAATGTTACCACCTTCCAAATGGGTTGCCAGATTTCGATGGGCAAGAAAGAGCCTGTTCAATTTGGAGAAGATATGCTAATTTGTTCGAGACCTCTCAGTCGCTGTTCTGGGCTTCTTTTGGGCTCGTCGATTTGACGACGTTCGAGTTGACCGGTATCAAAAGTTTTACAAGATTTTGGGCTCTTCTGACTTTTGGATCGTATTCCGTTATTAACATAATAGTCCTGCTCAACATGCTGATTGCCATGATGTCGAACTCATATCAAATAATTTCCGAACGAGCGGATACTGAATGGAAATTCGCCCGCAGTAATCTTTGGATGTCTTACTTCGAAGATGGCGACACCGTCCCGCCGCCGTTCAACATCATACCGACGCCAAAGCATTTCTTTTGCTGGATCAAAAGCTATTTTAACAAACGAGAAAGGCGTTCAATACTCAACAAATCTCGTGAAAAAGCTAGACAGGAACACGAAGCGGTAATGAGGATATTAGTCAGAAGATACGTGACAGCAGAACAAAGGGCTAGAGATGAAGTAGGAGTCACGGAGGATGACGTTATGGAGATCCGTCAAGATATATCCACACTCAGATACGAGCTCATTGATATACTGCATAACAATGGAATGAAGACACCAAGAGTCAGCTTGCAGGATACTGCCGGTAATAATTATAATCTTAAAATTTCGAATTATCTACACTACCCGATGATTGTTTAA
Protein
MWYYAELEKDKCYHLPNGLPDFDGQERACSIWRRYANLFETSQSLFWASFGLVDLTTFELTGIKSFTRFWALLTFGSYSVINIIVLLNMLIAMMSNSYQIISERADTEWKFARSNLWMSYFEDGDTVPPPFNIIPTPKHFFCWIKSYFNKRERRSILNKSREKARQEHEAVMRILVRRYVTAEQRARDEVGVTEDDVMEIRQDISTLRYELIDILHNNGMKTPRVSLQDTAGNNYNLKISNYLHYPMIV
Summary
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Uniprot
H9JIC2
A0A2H1VJJ5
A0A2H1VZ28
A0A2A4J1C2
A0A2A4J032
A0A2W1BK16
+ More
A0A437BN04 A0A194Q499 A0A212EM62 A0A1B6EEM2 A0A1B6D2D8 A0A0C9R7U6 A0A0C9QH65 U4UUV8 A0A0J7NDJ6 A0A182XBR3 K7J2P6 N6TRC9 A0A0N0U7F9 N6TWB0 A0A2J7PZ35 A0A182UDG9 Q17A16 A0A1S4FAK9 A0A182RQ80 A0A182FIQ9 A0A182LT00 A0A182YK11 A0A182N0R8 A0A182GH52 A0A182T490 A0A1S4G9C1 A0A182KCS2 A0A182PV03 Q7PS36 A0A182HMZ4 A0A182QF93 A0A182W588 A0A084VN48 A0A336MTI6 A0A154PFZ3 A0A2A3E6M6 A0A182JIJ2 A0A087ZR90 A0A232EFJ4 A0A3L8D5X4 A0A026VYU6 B0XHC6 A0A1W4WU54 E2BPR3 A0A1Q3FSE9 A0A2P8Y1V1 A0A1B6FTI6 E1ZZG0 A0A336MBJ8 A0A146KZM0 A0A151XBS4 A0A195FV57 A0A158NY16 M4N9S2 A0A1B6LAJ0 F4WNP5 A0A0B4VRY1 A0A195B7N8 A0A310SA84 A0A195EC27 A0A384THF3 A0A1J1HYQ2 A0A139WIM8 A0A2S2QNH1 A0A0L7KHH4 A0A2H8TTI6 A0A1I8NT53 A0A1B0BW76 A0A1A9XVK1 A0A1A9WYZ7 A0A1B0GFC1 A0A1B0AJS8 A0A1I8PGE5 Q94447 A0A3B0K5L0 A0A1I8MDK6 A0A1A9VFI0 T1P894 A0A0M4ERZ4 A0A0L0CIC2 B4M5X7 A0A1W4UPV3 B4JU24 F0JAI0 A0A2H8TML4 B4K6J2 B4GNE7 Q29C99 B3P828 A0A0A1X868 A0A3B0JEU9 J9KVL3 B4HZH7 U3PXB8
A0A437BN04 A0A194Q499 A0A212EM62 A0A1B6EEM2 A0A1B6D2D8 A0A0C9R7U6 A0A0C9QH65 U4UUV8 A0A0J7NDJ6 A0A182XBR3 K7J2P6 N6TRC9 A0A0N0U7F9 N6TWB0 A0A2J7PZ35 A0A182UDG9 Q17A16 A0A1S4FAK9 A0A182RQ80 A0A182FIQ9 A0A182LT00 A0A182YK11 A0A182N0R8 A0A182GH52 A0A182T490 A0A1S4G9C1 A0A182KCS2 A0A182PV03 Q7PS36 A0A182HMZ4 A0A182QF93 A0A182W588 A0A084VN48 A0A336MTI6 A0A154PFZ3 A0A2A3E6M6 A0A182JIJ2 A0A087ZR90 A0A232EFJ4 A0A3L8D5X4 A0A026VYU6 B0XHC6 A0A1W4WU54 E2BPR3 A0A1Q3FSE9 A0A2P8Y1V1 A0A1B6FTI6 E1ZZG0 A0A336MBJ8 A0A146KZM0 A0A151XBS4 A0A195FV57 A0A158NY16 M4N9S2 A0A1B6LAJ0 F4WNP5 A0A0B4VRY1 A0A195B7N8 A0A310SA84 A0A195EC27 A0A384THF3 A0A1J1HYQ2 A0A139WIM8 A0A2S2QNH1 A0A0L7KHH4 A0A2H8TTI6 A0A1I8NT53 A0A1B0BW76 A0A1A9XVK1 A0A1A9WYZ7 A0A1B0GFC1 A0A1B0AJS8 A0A1I8PGE5 Q94447 A0A3B0K5L0 A0A1I8MDK6 A0A1A9VFI0 T1P894 A0A0M4ERZ4 A0A0L0CIC2 B4M5X7 A0A1W4UPV3 B4JU24 F0JAI0 A0A2H8TML4 B4K6J2 B4GNE7 Q29C99 B3P828 A0A0A1X868 A0A3B0JEU9 J9KVL3 B4HZH7 U3PXB8
Pubmed
EMBL
BABH01022307
BABH01022308
ODYU01002911
SOQ40997.1
ODYU01005013
SOQ45484.1
+ More
NWSH01004304 PCG65240.1 PCG65239.1 KZ150121 PZC73176.1 RSAL01000028 RVE51850.1 KQ459585 KPI98235.1 AGBW02013881 OWR42590.1 GEDC01000920 JAS36378.1 GEDC01017419 JAS19879.1 GBYB01002831 JAG72598.1 GBYB01002829 JAG72596.1 KB632375 ERL93960.1 LBMM01006483 KMQ90580.1 APGK01007747 APGK01007748 APGK01007749 APGK01007750 KB737435 ENN83024.1 KQ435699 KOX80578.1 APGK01013991 APGK01013992 APGK01013993 APGK01013994 APGK01013995 APGK01013996 KB739168 ENN82358.1 NEVH01020341 PNF21582.1 CH477341 EAT43090.1 AXCM01000046 JXUM01063153 JXUM01063154 KQ562234 KXJ76346.1 AAAB01008846 EAA06236.6 APCN01004943 AXCN02001396 AXCN02001397 ATLV01014688 ATLV01014689 KE524978 KFB39392.1 UFQT01002598 SSX33742.1 KQ434896 KZC10761.1 KZ288375 PBC26701.1 NNAY01004990 OXU17130.1 QOIP01000012 RLU15644.1 KK107558 EZA48978.1 DS233135 EDS28306.1 GL449658 EFN82264.1 GFDL01004514 JAV30531.1 PYGN01001038 PSN38237.1 GECZ01016223 JAS53546.1 GL435343 EFN73452.1 UFQT01000867 SSX27732.1 GDHC01017644 JAQ00985.1 KQ982316 KYQ57836.1 KQ981215 KYN44525.1 ADTU01002882 ADTU01002883 ADTU01002884 KC329816 AGG86916.1 GEBQ01019250 JAT20727.1 GL888237 EGI64238.1 KM280572 AJD25385.1 KQ976565 KYM80531.1 KQ765605 OAD53838.1 KQ979074 KYN22778.1 KX249688 AOR81465.1 CVRI01000020 CRK91249.1 KQ971338 KYB27779.1 GGMS01009469 MBY78672.1 JTDY01009830 KOB62560.1 GFXV01005326 MBW17131.1 JXJN01021660 JXJN01021661 CCAG010009340 Z80230 CAB02410.1 OUUW01000005 SPP80906.1 KA644906 AFP59535.1 CP012526 ALC48014.1 JRES01000338 KNC32158.1 CH940652 EDW59053.1 CH916374 EDV90994.1 BT126022 ADY17721.1 GFXV01003366 MBW15171.1 CH933806 EDW16292.1 CH479186 EDW39273.1 CM000070 EAL26747.2 CH954182 EDV53155.1 GBXI01007439 JAD06853.1 SPP80904.1 ABLF02038759 ABLF02038765 ABLF02038771 CH480819 EDW53434.1 BT150384 AGW52194.1
NWSH01004304 PCG65240.1 PCG65239.1 KZ150121 PZC73176.1 RSAL01000028 RVE51850.1 KQ459585 KPI98235.1 AGBW02013881 OWR42590.1 GEDC01000920 JAS36378.1 GEDC01017419 JAS19879.1 GBYB01002831 JAG72598.1 GBYB01002829 JAG72596.1 KB632375 ERL93960.1 LBMM01006483 KMQ90580.1 APGK01007747 APGK01007748 APGK01007749 APGK01007750 KB737435 ENN83024.1 KQ435699 KOX80578.1 APGK01013991 APGK01013992 APGK01013993 APGK01013994 APGK01013995 APGK01013996 KB739168 ENN82358.1 NEVH01020341 PNF21582.1 CH477341 EAT43090.1 AXCM01000046 JXUM01063153 JXUM01063154 KQ562234 KXJ76346.1 AAAB01008846 EAA06236.6 APCN01004943 AXCN02001396 AXCN02001397 ATLV01014688 ATLV01014689 KE524978 KFB39392.1 UFQT01002598 SSX33742.1 KQ434896 KZC10761.1 KZ288375 PBC26701.1 NNAY01004990 OXU17130.1 QOIP01000012 RLU15644.1 KK107558 EZA48978.1 DS233135 EDS28306.1 GL449658 EFN82264.1 GFDL01004514 JAV30531.1 PYGN01001038 PSN38237.1 GECZ01016223 JAS53546.1 GL435343 EFN73452.1 UFQT01000867 SSX27732.1 GDHC01017644 JAQ00985.1 KQ982316 KYQ57836.1 KQ981215 KYN44525.1 ADTU01002882 ADTU01002883 ADTU01002884 KC329816 AGG86916.1 GEBQ01019250 JAT20727.1 GL888237 EGI64238.1 KM280572 AJD25385.1 KQ976565 KYM80531.1 KQ765605 OAD53838.1 KQ979074 KYN22778.1 KX249688 AOR81465.1 CVRI01000020 CRK91249.1 KQ971338 KYB27779.1 GGMS01009469 MBY78672.1 JTDY01009830 KOB62560.1 GFXV01005326 MBW17131.1 JXJN01021660 JXJN01021661 CCAG010009340 Z80230 CAB02410.1 OUUW01000005 SPP80906.1 KA644906 AFP59535.1 CP012526 ALC48014.1 JRES01000338 KNC32158.1 CH940652 EDW59053.1 CH916374 EDV90994.1 BT126022 ADY17721.1 GFXV01003366 MBW15171.1 CH933806 EDW16292.1 CH479186 EDW39273.1 CM000070 EAL26747.2 CH954182 EDV53155.1 GBXI01007439 JAD06853.1 SPP80904.1 ABLF02038759 ABLF02038765 ABLF02038771 CH480819 EDW53434.1 BT150384 AGW52194.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000030742
+ More
UP000036403 UP000076407 UP000002358 UP000019118 UP000053105 UP000235965 UP000075902 UP000008820 UP000075900 UP000069272 UP000075883 UP000076408 UP000075884 UP000069940 UP000249989 UP000075901 UP000075881 UP000075885 UP000007062 UP000075840 UP000075886 UP000075920 UP000030765 UP000076502 UP000242457 UP000075880 UP000005203 UP000215335 UP000279307 UP000053097 UP000002320 UP000192223 UP000008237 UP000245037 UP000000311 UP000075809 UP000078541 UP000005205 UP000007755 UP000078540 UP000078492 UP000183832 UP000007266 UP000037510 UP000095300 UP000092460 UP000092443 UP000091820 UP000092444 UP000092445 UP000268350 UP000095301 UP000078200 UP000092553 UP000037069 UP000008792 UP000192221 UP000001070 UP000009192 UP000008744 UP000001819 UP000008711 UP000007819 UP000001292
UP000036403 UP000076407 UP000002358 UP000019118 UP000053105 UP000235965 UP000075902 UP000008820 UP000075900 UP000069272 UP000075883 UP000076408 UP000075884 UP000069940 UP000249989 UP000075901 UP000075881 UP000075885 UP000007062 UP000075840 UP000075886 UP000075920 UP000030765 UP000076502 UP000242457 UP000075880 UP000005203 UP000215335 UP000279307 UP000053097 UP000002320 UP000192223 UP000008237 UP000245037 UP000000311 UP000075809 UP000078541 UP000005205 UP000007755 UP000078540 UP000078492 UP000183832 UP000007266 UP000037510 UP000095300 UP000092460 UP000092443 UP000091820 UP000092444 UP000092445 UP000268350 UP000095301 UP000078200 UP000092553 UP000037069 UP000008792 UP000192221 UP000001070 UP000009192 UP000008744 UP000001819 UP000008711 UP000007819 UP000001292
Pfam
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
ProteinModelPortal
H9JIC2
A0A2H1VJJ5
A0A2H1VZ28
A0A2A4J1C2
A0A2A4J032
A0A2W1BK16
+ More
A0A437BN04 A0A194Q499 A0A212EM62 A0A1B6EEM2 A0A1B6D2D8 A0A0C9R7U6 A0A0C9QH65 U4UUV8 A0A0J7NDJ6 A0A182XBR3 K7J2P6 N6TRC9 A0A0N0U7F9 N6TWB0 A0A2J7PZ35 A0A182UDG9 Q17A16 A0A1S4FAK9 A0A182RQ80 A0A182FIQ9 A0A182LT00 A0A182YK11 A0A182N0R8 A0A182GH52 A0A182T490 A0A1S4G9C1 A0A182KCS2 A0A182PV03 Q7PS36 A0A182HMZ4 A0A182QF93 A0A182W588 A0A084VN48 A0A336MTI6 A0A154PFZ3 A0A2A3E6M6 A0A182JIJ2 A0A087ZR90 A0A232EFJ4 A0A3L8D5X4 A0A026VYU6 B0XHC6 A0A1W4WU54 E2BPR3 A0A1Q3FSE9 A0A2P8Y1V1 A0A1B6FTI6 E1ZZG0 A0A336MBJ8 A0A146KZM0 A0A151XBS4 A0A195FV57 A0A158NY16 M4N9S2 A0A1B6LAJ0 F4WNP5 A0A0B4VRY1 A0A195B7N8 A0A310SA84 A0A195EC27 A0A384THF3 A0A1J1HYQ2 A0A139WIM8 A0A2S2QNH1 A0A0L7KHH4 A0A2H8TTI6 A0A1I8NT53 A0A1B0BW76 A0A1A9XVK1 A0A1A9WYZ7 A0A1B0GFC1 A0A1B0AJS8 A0A1I8PGE5 Q94447 A0A3B0K5L0 A0A1I8MDK6 A0A1A9VFI0 T1P894 A0A0M4ERZ4 A0A0L0CIC2 B4M5X7 A0A1W4UPV3 B4JU24 F0JAI0 A0A2H8TML4 B4K6J2 B4GNE7 Q29C99 B3P828 A0A0A1X868 A0A3B0JEU9 J9KVL3 B4HZH7 U3PXB8
A0A437BN04 A0A194Q499 A0A212EM62 A0A1B6EEM2 A0A1B6D2D8 A0A0C9R7U6 A0A0C9QH65 U4UUV8 A0A0J7NDJ6 A0A182XBR3 K7J2P6 N6TRC9 A0A0N0U7F9 N6TWB0 A0A2J7PZ35 A0A182UDG9 Q17A16 A0A1S4FAK9 A0A182RQ80 A0A182FIQ9 A0A182LT00 A0A182YK11 A0A182N0R8 A0A182GH52 A0A182T490 A0A1S4G9C1 A0A182KCS2 A0A182PV03 Q7PS36 A0A182HMZ4 A0A182QF93 A0A182W588 A0A084VN48 A0A336MTI6 A0A154PFZ3 A0A2A3E6M6 A0A182JIJ2 A0A087ZR90 A0A232EFJ4 A0A3L8D5X4 A0A026VYU6 B0XHC6 A0A1W4WU54 E2BPR3 A0A1Q3FSE9 A0A2P8Y1V1 A0A1B6FTI6 E1ZZG0 A0A336MBJ8 A0A146KZM0 A0A151XBS4 A0A195FV57 A0A158NY16 M4N9S2 A0A1B6LAJ0 F4WNP5 A0A0B4VRY1 A0A195B7N8 A0A310SA84 A0A195EC27 A0A384THF3 A0A1J1HYQ2 A0A139WIM8 A0A2S2QNH1 A0A0L7KHH4 A0A2H8TTI6 A0A1I8NT53 A0A1B0BW76 A0A1A9XVK1 A0A1A9WYZ7 A0A1B0GFC1 A0A1B0AJS8 A0A1I8PGE5 Q94447 A0A3B0K5L0 A0A1I8MDK6 A0A1A9VFI0 T1P894 A0A0M4ERZ4 A0A0L0CIC2 B4M5X7 A0A1W4UPV3 B4JU24 F0JAI0 A0A2H8TML4 B4K6J2 B4GNE7 Q29C99 B3P828 A0A0A1X868 A0A3B0JEU9 J9KVL3 B4HZH7 U3PXB8
PDB
6G1K
E-value=2.20102e-39,
Score=405
Ontologies
GO
GO:0016021
GO:0005262
GO:0016020
GO:0070588
GO:0015279
GO:0051480
GO:0070679
GO:0005887
GO:0034703
GO:0006828
GO:0006811
GO:0007605
GO:0042803
GO:0046982
GO:0008355
GO:0010461
GO:0016027
GO:0008104
GO:0071454
GO:0050908
GO:0035997
GO:0007005
GO:0001895
GO:0008377
GO:0005216
GO:0005839
GO:0051603
GO:0030145
GO:0006412
GO:0016876
GO:0043039
GO:0055085
PANTHER
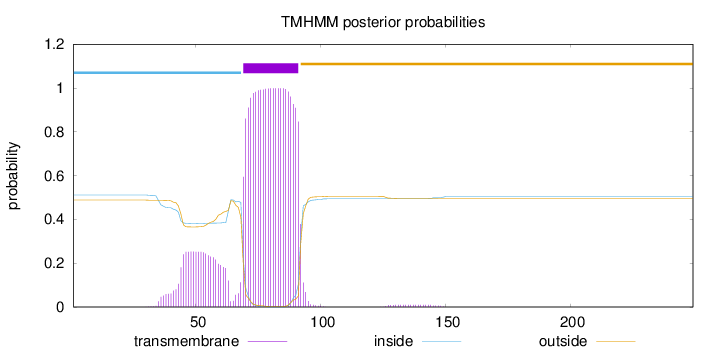
Topology
Length:
249
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
28.02129
Exp number, first 60 AAs:
4.55519
Total prob of N-in:
0.51181
inside
1 - 68
TMhelix
69 - 91
outside
92 - 249
Population Genetic Test Statistics
Pi
153.611061
Theta
122.623004
Tajima's D
0.754317
CLR
0.080592
CSRT
0.591420428978551
Interpretation
Uncertain
