Gene
KWMTBOMO08345
Pre Gene Modal
BGIBMGA009272
Annotation
PREDICTED:_transient_receptor_potential_protein_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.61 Extracellular Reliability : 1.975
Sequence
CDS
ATGAACCGCGGCTCCAGAGAAGAGCTCCTGGGTAGCTCCCAGGAAGCCCTGAGGAACAGTCAGCATGACCTTCGCTTTACTTGCGAGCGCCCTCTGACAAGAGTCGAGAAGACATATCTGCTGCATGCTGACAGAGGGGACTACGTCACAGTAAAAAGGTTGATTGAGCAGAACCTTGGAAAACCAGAGGTACTGGACATCAACTGTGTGGATCCACTCAATAGGTCGGCCCTCATCGCTGCAATTGAGAACGAGAATATTGAACTCATCAAACTACTTCTTGGATCGGGAATCAAAGTTAAGGATGCTCTGCTCCACGCTATCAAAGAAGAATATGTGGAAGCTGTAGAATTGCTGCTGCAATGGGAAGAGGAACATTATGAGCCTGGAGAACCTTATGAAGGGATAATGCTTCTATGCAAGACTGTCGAGAGAAATCAGCTATCTTGCCGATTAATATACTCGATTCCAATCTAA
Protein
MNRGSREELLGSSQEALRNSQHDLRFTCERPLTRVEKTYLLHADRGDYVTVKRLIEQNLGKPEVLDINCVDPLNRSALIAAIENENIELIKLLLGSGIKVKDALLHAIKEEYVEAVELLLQWEEEHYEPGEPYEGIMLLCKTVERNQLSCRLIYSIPI
Summary
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Uniprot
A0A2H1VJ93
A0A437BN04
A0A2A4J1C2
A0A2W1BK16
A0A2A4J032
A0A0N1PKC9
+ More
A0A194Q499 A0A212EM62 A0A1B6E6H8 A0A1B6DBK1 A0A0J7KK40 N6TWB0 U4UUV8 A0A1B6KIV2 A0A2H8TML4 A0A2H8TQN6 A0A384THF3 A0A1B6H849 E1ZZG0 J9KVL3 A0A0B4VRY1 A0A1B6FQL6 E2BPR3 A0A0C9QH65 M4N9S2 A0A0N0U7F9 A0A2P8Y1V1 A0A1S4EAA7 A0A2A3E6M6 A0A3L8D5X4 A0A146KZM0 A0A026VYU6 K7J2P6 A0A087ZR90 A0A2J7PZ24 A0A1D2MWP4 A0A0L0CIC2 A0A1J1HYQ2 E0VB86 A0A0N8B8M1 A0A0P5DRS4 A0A0P5BZ13 A0A0N8BB71 A0A0P5D6N5 B3LYU7 A0A0P6HSS9 A0A0P6ALR9 A0A0P5FVE1 A0A0P5D2G9 A0A1W4UPV3
A0A194Q499 A0A212EM62 A0A1B6E6H8 A0A1B6DBK1 A0A0J7KK40 N6TWB0 U4UUV8 A0A1B6KIV2 A0A2H8TML4 A0A2H8TQN6 A0A384THF3 A0A1B6H849 E1ZZG0 J9KVL3 A0A0B4VRY1 A0A1B6FQL6 E2BPR3 A0A0C9QH65 M4N9S2 A0A0N0U7F9 A0A2P8Y1V1 A0A1S4EAA7 A0A2A3E6M6 A0A3L8D5X4 A0A146KZM0 A0A026VYU6 K7J2P6 A0A087ZR90 A0A2J7PZ24 A0A1D2MWP4 A0A0L0CIC2 A0A1J1HYQ2 E0VB86 A0A0N8B8M1 A0A0P5DRS4 A0A0P5BZ13 A0A0N8BB71 A0A0P5D6N5 B3LYU7 A0A0P6HSS9 A0A0P6ALR9 A0A0P5FVE1 A0A0P5D2G9 A0A1W4UPV3
Pubmed
EMBL
ODYU01002858
SOQ40861.1
RSAL01000028
RVE51850.1
NWSH01004304
PCG65240.1
+ More
KZ150121 PZC73176.1 PCG65239.1 KQ459786 KPJ19923.1 KQ459585 KPI98235.1 AGBW02013881 OWR42590.1 GEDC01003799 JAS33499.1 GEDC01014250 JAS23048.1 LBMM01006483 KMQ90579.1 APGK01013991 APGK01013992 APGK01013993 APGK01013994 APGK01013995 APGK01013996 KB739168 ENN82358.1 KB632375 ERL93960.1 GEBQ01028595 JAT11382.1 GFXV01003366 MBW15171.1 GFXV01004504 MBW16309.1 KX249688 AOR81465.1 GECU01036923 JAS70783.1 GL435343 EFN73452.1 ABLF02038759 ABLF02038765 ABLF02038771 KM280572 AJD25385.1 GECZ01017264 JAS52505.1 GL449658 EFN82264.1 GBYB01002829 JAG72596.1 KC329816 AGG86916.1 KQ435699 KOX80578.1 PYGN01001038 PSN38237.1 KZ288375 PBC26701.1 QOIP01000012 RLU15644.1 GDHC01017644 JAQ00985.1 KK107558 EZA48978.1 NEVH01020341 PNF21594.1 LJIJ01000448 ODM97382.1 JRES01000338 KNC32158.1 CVRI01000020 CRK91249.1 AAZO01000650 DS235023 EEB10642.1 GDIQ01202091 JAK49634.1 GDIP01153685 JAJ69717.1 GDIP01177990 JAJ45412.1 GDIQ01194891 JAK56834.1 GDIP01165468 JAJ57934.1 CH902617 EDV44063.1 GDIQ01015497 JAN79240.1 GDIP01027505 JAM76210.1 GDIQ01250973 JAK00752.1 GDIP01177989 JAJ45413.1
KZ150121 PZC73176.1 PCG65239.1 KQ459786 KPJ19923.1 KQ459585 KPI98235.1 AGBW02013881 OWR42590.1 GEDC01003799 JAS33499.1 GEDC01014250 JAS23048.1 LBMM01006483 KMQ90579.1 APGK01013991 APGK01013992 APGK01013993 APGK01013994 APGK01013995 APGK01013996 KB739168 ENN82358.1 KB632375 ERL93960.1 GEBQ01028595 JAT11382.1 GFXV01003366 MBW15171.1 GFXV01004504 MBW16309.1 KX249688 AOR81465.1 GECU01036923 JAS70783.1 GL435343 EFN73452.1 ABLF02038759 ABLF02038765 ABLF02038771 KM280572 AJD25385.1 GECZ01017264 JAS52505.1 GL449658 EFN82264.1 GBYB01002829 JAG72596.1 KC329816 AGG86916.1 KQ435699 KOX80578.1 PYGN01001038 PSN38237.1 KZ288375 PBC26701.1 QOIP01000012 RLU15644.1 GDHC01017644 JAQ00985.1 KK107558 EZA48978.1 NEVH01020341 PNF21594.1 LJIJ01000448 ODM97382.1 JRES01000338 KNC32158.1 CVRI01000020 CRK91249.1 AAZO01000650 DS235023 EEB10642.1 GDIQ01202091 JAK49634.1 GDIP01153685 JAJ69717.1 GDIP01177990 JAJ45412.1 GDIQ01194891 JAK56834.1 GDIP01165468 JAJ57934.1 CH902617 EDV44063.1 GDIQ01015497 JAN79240.1 GDIP01027505 JAM76210.1 GDIQ01250973 JAK00752.1 GDIP01177989 JAJ45413.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
UP000036403
+ More
UP000019118 UP000030742 UP000000311 UP000007819 UP000008237 UP000053105 UP000245037 UP000079169 UP000242457 UP000279307 UP000053097 UP000002358 UP000005203 UP000235965 UP000094527 UP000037069 UP000183832 UP000009046 UP000007801 UP000192221
UP000019118 UP000030742 UP000000311 UP000007819 UP000008237 UP000053105 UP000245037 UP000079169 UP000242457 UP000279307 UP000053097 UP000002358 UP000005203 UP000235965 UP000094527 UP000037069 UP000183832 UP000009046 UP000007801 UP000192221
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
ProteinModelPortal
A0A2H1VJ93
A0A437BN04
A0A2A4J1C2
A0A2W1BK16
A0A2A4J032
A0A0N1PKC9
+ More
A0A194Q499 A0A212EM62 A0A1B6E6H8 A0A1B6DBK1 A0A0J7KK40 N6TWB0 U4UUV8 A0A1B6KIV2 A0A2H8TML4 A0A2H8TQN6 A0A384THF3 A0A1B6H849 E1ZZG0 J9KVL3 A0A0B4VRY1 A0A1B6FQL6 E2BPR3 A0A0C9QH65 M4N9S2 A0A0N0U7F9 A0A2P8Y1V1 A0A1S4EAA7 A0A2A3E6M6 A0A3L8D5X4 A0A146KZM0 A0A026VYU6 K7J2P6 A0A087ZR90 A0A2J7PZ24 A0A1D2MWP4 A0A0L0CIC2 A0A1J1HYQ2 E0VB86 A0A0N8B8M1 A0A0P5DRS4 A0A0P5BZ13 A0A0N8BB71 A0A0P5D6N5 B3LYU7 A0A0P6HSS9 A0A0P6ALR9 A0A0P5FVE1 A0A0P5D2G9 A0A1W4UPV3
A0A194Q499 A0A212EM62 A0A1B6E6H8 A0A1B6DBK1 A0A0J7KK40 N6TWB0 U4UUV8 A0A1B6KIV2 A0A2H8TML4 A0A2H8TQN6 A0A384THF3 A0A1B6H849 E1ZZG0 J9KVL3 A0A0B4VRY1 A0A1B6FQL6 E2BPR3 A0A0C9QH65 M4N9S2 A0A0N0U7F9 A0A2P8Y1V1 A0A1S4EAA7 A0A2A3E6M6 A0A3L8D5X4 A0A146KZM0 A0A026VYU6 K7J2P6 A0A087ZR90 A0A2J7PZ24 A0A1D2MWP4 A0A0L0CIC2 A0A1J1HYQ2 E0VB86 A0A0N8B8M1 A0A0P5DRS4 A0A0P5BZ13 A0A0N8BB71 A0A0P5D6N5 B3LYU7 A0A0P6HSS9 A0A0P6ALR9 A0A0P5FVE1 A0A0P5D2G9 A0A1W4UPV3
PDB
5Z96
E-value=3.09214e-09,
Score=142
Ontologies
GO
PANTHER
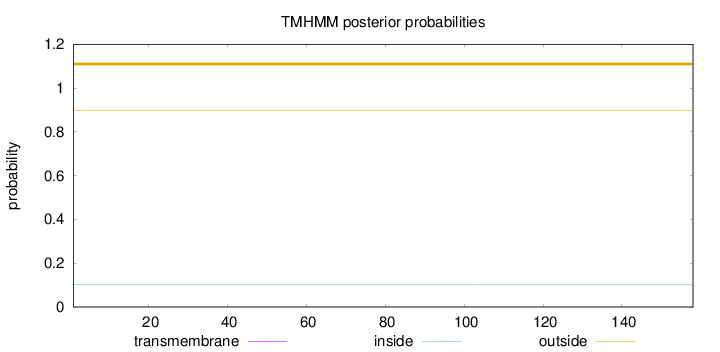
Topology
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2e-05
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10292
outside
1 - 158
Population Genetic Test Statistics
Pi
24.915172
Theta
180.327604
Tajima's D
0.577918
CLR
1.157217
CSRT
0.539823008849557
Interpretation
Uncertain
