Gene
KWMTBOMO08343 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009235
Annotation
PREDICTED:_alpha-N-acetylglucosaminidase_isoform_X1_[Bombyx_mori]
Full name
Proteasome endopeptidase complex
Location in the cell
PlasmaMembrane Reliability : 1.257
Sequence
CDS
ATGTTCAGGAACCAGTACGATAGCGACGTCACAGTATGGAGTCCTCAAGGCCGTCTCCATCAAGTGGAGTATGCAATGGAAGCTGTGAAACTTGGTTCTGCAACCATTGGTCTGAAGAATAAGGACTATGCGGTGTTGATAGCGTTGAAACGCGCTGTGAGCGAATTGTCCGCATATCAGAAGAAAATTATTCCTATTGATGAACATATTGGAATTTCTATATCAGGTCTTACTGCAGATGCTCGTATGCTGAGTCGCTACATGCGCACAGAGTGTCTCAATCACCGATATTCTCATGATGCTCCAATGCCCGTTGGACGTCTCATCTCGTCAGTTGGTAACAAAATGCAGATTTGTACCCAGAGATACGACAAGCGACCCTTGGGTGTGGGCTTGTTGGTTGCTGGTTATGATGACCAAGGACCTCATATTTACCAGACTTGTCCATCAGCCAACTACTTTGATTGTCGTGCGATGGCAATCGGAGCACGTTCGCAGTCAGCGCGTACTTATCTTGAGAAGCATCTCAATACATTCATGGATTGTGACCTTCAAGAGCTAGTAGCTCACGGTCTTAGAGCCCTACGGGATACGCTGCCAAATGAAGTTGATCTAAACACAAAGAATGTTTCTATATGTATCGTCGGTCCCAAGACTCCGTTGCGTATATGCGACGAGGAAGAGTTATCTCGACTGTTAGCTCTGGTCGAGGGTGAAGAGAGACGCGGCGGCACCGCGACCGGCGACGCGGGCGCTCCGGGAGGCGCCGCCCCGCGCGACCAGGGGGACGACCGGGACCCACAAGCGGCGAGTCAAATAGCGTGGCAAGTTCAACGAGTTGTGGTGCCGGAACCGCTCCCAGAAGTCGACGAACTTGTCGTTTCTAATGATAGATTTCGCTACTATCAAAACGTGTGCACGGCGTCTTATTCATTCGTTTGGTGGCAGACAAACGATTGGGTCGAACACGTCCAATGGATGGCACTGAATGGTATTAACTTAGCTTTAGCGCCAGTTGCACAGGAAGCGGCTTGGGCTCGCGTCTATCGCAGCCTCGGAATGACGGACGACGAAATAGATGAGCATTTTACCGGTCCCGCGTTTTTGGCTTGGCTTCGTATGGGGAATGTACACGGCTGGGGTGGACCACTCCTCAAGTCATGGAACGACAAACAAAGAGAAATTCAAGATTCCGTAATCGACTACATGTACCAGCTTGGCATTGTATCGGTCTTCCCGGCTTTCAATGGTCACGTACCGAAGGCATTTGCACGAATATTTCCGAATGTAACTTTTCACGACGTCGATACGTGGAATAAATTTGACGCTGAACATTGTTGTGGCTTGTTCGTAGATCCGAATGAACCACTTTTTAAAATAATAGGGAAAATGTTTTTAAAAGAAACTGGCAGTTTGTCAGCCACAAGTCACATTTACTCTTCGGACCCATTTAACGAGGTTAAAATTGAACAATGGTCAACAGCTTTGGTTGTGCAAACGGCCAAATCTATATTCTCGACATTATTAGAATTCGATGACGAAGCTGTCTGGTTATTGCAGAATTGGATGTTTGTTAGCGATCCGTTACAATGGCCTAAATCTAGGGTACGGTCATTTATGACGTCAGTGCCGAATGGTAGAATGTTGGTACTGGACCTTCAAGCGGAGCAGTGGCCACAATACGAACTTTACGAGATGTATTTCGGTCAACCATTCATATGGTGCATGTTACACAATTTTGGCGGAACTTTAGGGATGTTTGGTAACATGATAACGATCAATAAAGAAGTGTATTATGCACGGGCCCTCGCAAATAGCACAATGATTGGCGTCGGCCTTACCCCTGAAGGAATAAATCAGAATTATGTAGTTTACGATTTAATGTTGGAGTCAGCTTGGCGTAAGAAACCCGTAGAAAATCTCGATACTTGGGCAGGAGAATACGCGGAGCGAAGGTACGGTTGCAACACGACAACGGACGCCTGGCGGTATCTGTTAAAAAGCGTATACAGCTTCAATGGCTTGAACAAAGTCCGAGGGAAGTACGTAATCACTCGTAGGCCCAGTTTCAACATTAAGCCATGGGCTTGGTACAAAAGTTACGATCTATTCCAGGCTTTGAAAAAGTTTGTCACGAGTACAGATTGTTACTCCGATGGATTTCTATATGATATTGTTGATGTTACAAGGCAAGCGTTACAGTACAGGGCGGAACAACTATACGTAAATATGCAGAGCGCACGCTATTCCAACACTTTGTTGTTCAAAGAAGCGATACAACAATTTCTGGATGCCATGATTGATATAGAATCGATATTGCGGACAAATACGTTTTTCGGTGCAGCCGATTGGATCGCTAAAGCTGAGAATTTCGCGAGAACACCAATAGAATCGTATTCATATAGTTTGAATGCTCGCAATCAAATAACTCTGTGGGGCCCTAACGGGGAAATAACAGATTATGCATGCAAACAATGGGCTGAGCTATTCCACTATTATTACATTCCTAGATGGACAAAGTTCTTGGATGTTGCCCTCGATGCCAAAACGAAACGCGAAGTCTTCGACGAGAAAACTGCTCGGGATATGATTCGATCCACAGTCGAGTATAAATTTCTGTTTGTGACTTTCGACACTATCCATCGACCTGCGGTTAACCCGATAGATCTTGCTCTGGATCTGTACCAAAAATGGGCGTTTTATCCTGGTCTCGATGATCTGCCGCAAAACGTTATCCGCCCCGATCCTGGAAAGAGAACCACCCTGCCCGACGTTGATAAAGAAAGCGATGAGACAGAAGATCCCCCTTCCGTTATTATGTGGCACTCCACAACGCCGATCAATTAG
Protein
MFRNQYDSDVTVWSPQGRLHQVEYAMEAVKLGSATIGLKNKDYAVLIALKRAVSELSAYQKKIIPIDEHIGISISGLTADARMLSRYMRTECLNHRYSHDAPMPVGRLISSVGNKMQICTQRYDKRPLGVGLLVAGYDDQGPHIYQTCPSANYFDCRAMAIGARSQSARTYLEKHLNTFMDCDLQELVAHGLRALRDTLPNEVDLNTKNVSICIVGPKTPLRICDEEELSRLLALVEGEERRGGTATGDAGAPGGAAPRDQGDDRDPQAASQIAWQVQRVVVPEPLPEVDELVVSNDRFRYYQNVCTASYSFVWWQTNDWVEHVQWMALNGINLALAPVAQEAAWARVYRSLGMTDDEIDEHFTGPAFLAWLRMGNVHGWGGPLLKSWNDKQREIQDSVIDYMYQLGIVSVFPAFNGHVPKAFARIFPNVTFHDVDTWNKFDAEHCCGLFVDPNEPLFKIIGKMFLKETGSLSATSHIYSSDPFNEVKIEQWSTALVVQTAKSIFSTLLEFDDEAVWLLQNWMFVSDPLQWPKSRVRSFMTSVPNGRMLVLDLQAEQWPQYELYEMYFGQPFIWCMLHNFGGTLGMFGNMITINKEVYYARALANSTMIGVGLTPEGINQNYVVYDLMLESAWRKKPVENLDTWAGEYAERRYGCNTTTDAWRYLLKSVYSFNGLNKVRGKYVITRRPSFNIKPWAWYKSYDLFQALKKFVTSTDCYSDGFLYDIVDVTRQALQYRAEQLYVNMQSARYSNTLLFKEAIQQFLDAMIDIESILRTNTFFGAADWIAKAENFARTPIESYSYSLNARNQITLWGPNGEITDYACKQWAELFHYYYIPRWTKFLDVALDAKTKREVFDEKTARDMIRSTVEYKFLFVTFDTIHRPAVNPIDLALDLYQKWAFYPGLDDLPQNVIRPDPGKRTTLPDVDKESDETEDPPSVIMWHSTTPIN
Summary
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Similarity
Belongs to the peptidase T1A family.
Uniprot
EC Number
3.4.25.1
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
PDB
4XWH
E-value=1.47494e-137,
Score=1258
Ontologies
GO
PANTHER
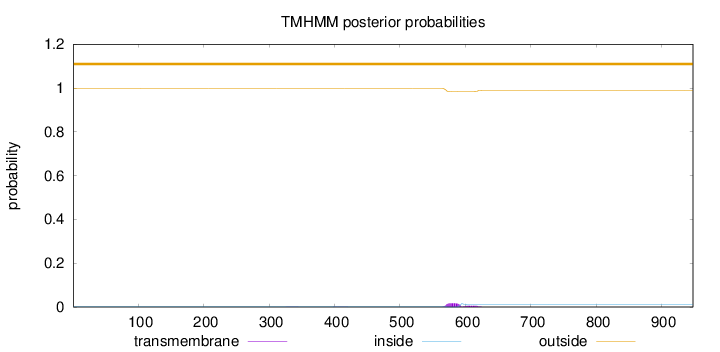
Topology
Subcellular location
Nucleus
Length:
948
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.501479999999999
Exp number, first 60 AAs:
0.00046
Total prob of N-in:
0.00121
outside
1 - 948
Population Genetic Test Statistics
Pi
220.265043
Theta
180.395712
Tajima's D
0.601653
CLR
0.459335
CSRT
0.548272586370681
Interpretation
Uncertain
