Gene
KWMTBOMO08328
Pre Gene Modal
BGIBMGA009265
Annotation
PREDICTED:_nuclear_transcription_factor_Y_subunit_alpha_[Amyelois_transitella]
Transcription factor
Location in the cell
Nuclear Reliability : 4.668
Sequence
CDS
ATGGAGCATGCACAAATCCAGAATGCAGATGGGCCCCAAGTGATGACCATCAATGGACAACAGTTGCAAGTATTGCAGATGAACACCTCTCCGCACATGGTTCAAGGTCCTAATGGTCAACAGATCATGGTTCATACAATAGCAACTCCTACACAGACTATTCAGGTAGCAGCTCCGAATGGGCAGCAGATCCAACAACTTCAAGTCTTACCGATTTCAAATTTGCAAGGTGGAGGGAACAATGTACAGCCAATGCAGTTAGTTCAGACACCTGACGGCCAGACATTTCTGTACCAACCGACTGCACAGGCTCAACCATCCATTGACCAACACCAAATAATTCATCAACCTGCCGTTATCAACTTGAATGGGAATCTTGTGCAAGTTGCTGGAGTGAGTAACGCCGTGCAGCCACAGCAGATCGTCAATCAACCTAATATTGTCATGATGGTGAATGGCACTAATAACACAAGCACAACAGCAACTCCAACAGAAGGTCCGGGATCATCAGCCGCTGGCTCTGATGAAGAGCCTCTGTTGTATGTAAATGCTAGACAGTTCAAGCAAATATTAAAACGTAGAGCTGCAAGAGCGAGGCTTCATGAACTAGGAAAAATACCTAAAGAACGGCCGAAATACCTGCACGAATCGCGACACAGGCACGCGATGAACAGAATCAGAGGTGAAGGTGGAAGGTTTCACTCGGGGAGTAGGAAAAATATGGACAATTCAGACCAGTCACATCTTTCAGAAGATCAGAAAGATGATATTAAACCGGACACTGTCTCCATCACAATCATACAGGAAGATGAAACTCCAGAACAACAGCAGACGCAGTGGAGACGGCTGGCGCCGCACCAGCCGCTACAGTCCACATAG
Protein
MEHAQIQNADGPQVMTINGQQLQVLQMNTSPHMVQGPNGQQIMVHTIATPTQTIQVAAPNGQQIQQLQVLPISNLQGGGNNVQPMQLVQTPDGQTFLYQPTAQAQPSIDQHQIIHQPAVINLNGNLVQVAGVSNAVQPQQIVNQPNIVMMVNGTNNTSTTATPTEGPGSSAAGSDEEPLLYVNARQFKQILKRRAARARLHELGKIPKERPKYLHESRHRHAMNRIRGEGGRFHSGSRKNMDNSDQSHLSEDQKDDIKPDTVSITIIQEDETPEQQQTQWRRLAPHQPLQST
Summary
Uniprot
H9JIB5
A0A3S2L9Q0
A0A2W1BUC1
A0A194PZP9
A0A2H1UZM1
A0A1E1WK50
+ More
S4P253 A0A212FDX3 A0A2A4IYZ3 A0A194R7J7 A0A1W4WP73 A0A1W4WP55 A0A1Y1MZN7 A0A1Y1MWE3 A0A1Y1MUU2 A0A1Y1MV75 A0A1Y1MUT0 A0A026WS19 A0A1B6DGI9 E0V9G5 D1ZZH5 A0A3L8DXG6 A0A2R7VPE6 A0A1B6LAP2 E9ISS0 A0A2J7PYW9 A0A2J7PYZ3 A0A088AGX0 A0A151IH48 A0A2J7PYZ1 A0A158NPE3 A0A0L7QK28 A0A195F4F0 A0A0A9ZII9 A0A224XTW4 A0A195E5B4 F4WH76 A0A2A3ESC0 A0A195B9D0 A0A023F394 E2ADU8 A0A154P6Q6 A0A0V0GCB0 A0A151X644 A0A0N0BFU8 A0A226E6R1 A0A1B6JM49 A0A0C9PJQ9 A0A1L8DHN4 A0A444TE91 A0A443QII4 A0A2D0PLM0 A0A131XG30 A0A131YQM8 A0A443SJD6 A0A2D0PJ21 A0A2D0PJ18 A0A224YUZ2 A0A2H8THC0 G1KSQ3 A0A1Q3F594 A0A1Q3F4F3 A0A0P4WE93 A0A3S2M950 A0A3P9H587 A0A3Q2XFX0 A0A2D0PLM6 A0A402EK00 B0R049 A0A3Q1FZ19 J9LW24 A0A315W4Z2 A0A1E1XDW9 A0A3Q3LQW8 A0A3B4WP72 A0A1A8HIU3 A0A1A8P5W2 A0A1A7ZBK8 A0A3Q3G304 A0A151P519 A0A1A7XWH7 V8P2H7 A0A2G8K6E2
S4P253 A0A212FDX3 A0A2A4IYZ3 A0A194R7J7 A0A1W4WP73 A0A1W4WP55 A0A1Y1MZN7 A0A1Y1MWE3 A0A1Y1MUU2 A0A1Y1MV75 A0A1Y1MUT0 A0A026WS19 A0A1B6DGI9 E0V9G5 D1ZZH5 A0A3L8DXG6 A0A2R7VPE6 A0A1B6LAP2 E9ISS0 A0A2J7PYW9 A0A2J7PYZ3 A0A088AGX0 A0A151IH48 A0A2J7PYZ1 A0A158NPE3 A0A0L7QK28 A0A195F4F0 A0A0A9ZII9 A0A224XTW4 A0A195E5B4 F4WH76 A0A2A3ESC0 A0A195B9D0 A0A023F394 E2ADU8 A0A154P6Q6 A0A0V0GCB0 A0A151X644 A0A0N0BFU8 A0A226E6R1 A0A1B6JM49 A0A0C9PJQ9 A0A1L8DHN4 A0A444TE91 A0A443QII4 A0A2D0PLM0 A0A131XG30 A0A131YQM8 A0A443SJD6 A0A2D0PJ21 A0A2D0PJ18 A0A224YUZ2 A0A2H8THC0 G1KSQ3 A0A1Q3F594 A0A1Q3F4F3 A0A0P4WE93 A0A3S2M950 A0A3P9H587 A0A3Q2XFX0 A0A2D0PLM6 A0A402EK00 B0R049 A0A3Q1FZ19 J9LW24 A0A315W4Z2 A0A1E1XDW9 A0A3Q3LQW8 A0A3B4WP72 A0A1A8HIU3 A0A1A8P5W2 A0A1A7ZBK8 A0A3Q3G304 A0A151P519 A0A1A7XWH7 V8P2H7 A0A2G8K6E2
Pubmed
EMBL
BABH01022259
RSAL01000371
RVE42139.1
KZ149916
PZC77951.1
KQ459585
+ More
KPI98219.1 ODYU01000005 SOQ34053.1 GDQN01005240 GDQN01003813 JAT85814.1 JAT87241.1 GAIX01012190 JAA80370.1 AGBW02008992 OWR51956.1 NWSH01005041 PCG64484.1 KQ460627 KPJ13489.1 GEZM01020031 GEZM01020030 JAV89446.1 GEZM01020029 GEZM01020028 JAV89448.1 GEZM01020034 GEZM01020032 JAV89442.1 GEZM01020027 JAV89449.1 GEZM01020033 JAV89443.1 KK107139 EZA57894.1 GEDC01019968 GEDC01019104 GEDC01012536 JAS17330.1 JAS18194.1 JAS24762.1 DS234993 EEB10021.1 KQ971338 EFA01850.1 QOIP01000003 RLU25164.1 KK854015 PTY09404.1 GEBQ01030102 GEBQ01019286 JAT09875.1 JAT20691.1 GL765434 EFZ16360.1 NEVH01020341 PNF21533.1 PNF21532.1 KQ977643 KYN00867.1 PNF21530.1 ADTU01022405 KQ415028 KOC58915.1 KQ981836 KYN35052.1 GBHO01000801 GBHO01000799 GBRD01014024 GDHC01011515 JAG42803.1 JAG42805.1 JAG51802.1 JAQ07114.1 GFTR01004883 JAW11543.1 KQ979608 KYN20276.1 GL888153 EGI66442.1 KZ288189 PBC34655.1 KQ976542 KYM81141.1 GBBI01003051 JAC15661.1 GL438820 EFN68460.1 KQ434827 KZC07557.1 GECL01000504 JAP05620.1 KQ982482 KYQ55851.1 KQ435794 KOX73868.1 LNIX01000006 OXA53295.1 GECU01016426 GECU01007743 JAS91280.1 JAS99963.1 GBYB01001193 JAG70960.1 GFDF01008163 JAV05921.1 SAUD01003300 RXG68324.1 NCKU01007194 RWS02819.1 GEFH01002478 JAP66103.1 GEDV01007715 JAP80842.1 NCKV01001865 RWS27639.1 GFPF01010281 MAA21427.1 GFXV01001711 MBW13516.1 GFDL01012310 JAV22735.1 GFDL01012605 JAV22440.1 GDRN01043654 JAI66973.1 CM012443 RVE70418.1 BDOT01000016 GCF44491.1 AL773601 CAQ14846.1 ABLF02038121 NHOQ01000318 PWA30927.1 GFAC01001726 JAT97462.1 HAEC01015934 SBQ84155.1 HAEG01006434 SBR76710.1 HADY01001216 SBP39701.1 AKHW03000979 KYO44019.1 HADW01008483 HADW01012833 HADX01000151 SBP22383.1 AZIM01001009 ETE68480.1 MRZV01000840 PIK43586.1
KPI98219.1 ODYU01000005 SOQ34053.1 GDQN01005240 GDQN01003813 JAT85814.1 JAT87241.1 GAIX01012190 JAA80370.1 AGBW02008992 OWR51956.1 NWSH01005041 PCG64484.1 KQ460627 KPJ13489.1 GEZM01020031 GEZM01020030 JAV89446.1 GEZM01020029 GEZM01020028 JAV89448.1 GEZM01020034 GEZM01020032 JAV89442.1 GEZM01020027 JAV89449.1 GEZM01020033 JAV89443.1 KK107139 EZA57894.1 GEDC01019968 GEDC01019104 GEDC01012536 JAS17330.1 JAS18194.1 JAS24762.1 DS234993 EEB10021.1 KQ971338 EFA01850.1 QOIP01000003 RLU25164.1 KK854015 PTY09404.1 GEBQ01030102 GEBQ01019286 JAT09875.1 JAT20691.1 GL765434 EFZ16360.1 NEVH01020341 PNF21533.1 PNF21532.1 KQ977643 KYN00867.1 PNF21530.1 ADTU01022405 KQ415028 KOC58915.1 KQ981836 KYN35052.1 GBHO01000801 GBHO01000799 GBRD01014024 GDHC01011515 JAG42803.1 JAG42805.1 JAG51802.1 JAQ07114.1 GFTR01004883 JAW11543.1 KQ979608 KYN20276.1 GL888153 EGI66442.1 KZ288189 PBC34655.1 KQ976542 KYM81141.1 GBBI01003051 JAC15661.1 GL438820 EFN68460.1 KQ434827 KZC07557.1 GECL01000504 JAP05620.1 KQ982482 KYQ55851.1 KQ435794 KOX73868.1 LNIX01000006 OXA53295.1 GECU01016426 GECU01007743 JAS91280.1 JAS99963.1 GBYB01001193 JAG70960.1 GFDF01008163 JAV05921.1 SAUD01003300 RXG68324.1 NCKU01007194 RWS02819.1 GEFH01002478 JAP66103.1 GEDV01007715 JAP80842.1 NCKV01001865 RWS27639.1 GFPF01010281 MAA21427.1 GFXV01001711 MBW13516.1 GFDL01012310 JAV22735.1 GFDL01012605 JAV22440.1 GDRN01043654 JAI66973.1 CM012443 RVE70418.1 BDOT01000016 GCF44491.1 AL773601 CAQ14846.1 ABLF02038121 NHOQ01000318 PWA30927.1 GFAC01001726 JAT97462.1 HAEC01015934 SBQ84155.1 HAEG01006434 SBR76710.1 HADY01001216 SBP39701.1 AKHW03000979 KYO44019.1 HADW01008483 HADW01012833 HADX01000151 SBP22383.1 AZIM01001009 ETE68480.1 MRZV01000840 PIK43586.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000218220
UP000053240
+ More
UP000192223 UP000053097 UP000009046 UP000007266 UP000279307 UP000235965 UP000005203 UP000078542 UP000005205 UP000053825 UP000078541 UP000078492 UP000007755 UP000242457 UP000078540 UP000000311 UP000076502 UP000075809 UP000053105 UP000198287 UP000288706 UP000285301 UP000221080 UP000288716 UP000001646 UP000265200 UP000264820 UP000288954 UP000257200 UP000007819 UP000261640 UP000261360 UP000261660 UP000050525 UP000230750
UP000192223 UP000053097 UP000009046 UP000007266 UP000279307 UP000235965 UP000005203 UP000078542 UP000005205 UP000053825 UP000078541 UP000078492 UP000007755 UP000242457 UP000078540 UP000000311 UP000076502 UP000075809 UP000053105 UP000198287 UP000288706 UP000285301 UP000221080 UP000288716 UP000001646 UP000265200 UP000264820 UP000288954 UP000257200 UP000007819 UP000261640 UP000261360 UP000261660 UP000050525 UP000230750
PRIDE
ProteinModelPortal
H9JIB5
A0A3S2L9Q0
A0A2W1BUC1
A0A194PZP9
A0A2H1UZM1
A0A1E1WK50
+ More
S4P253 A0A212FDX3 A0A2A4IYZ3 A0A194R7J7 A0A1W4WP73 A0A1W4WP55 A0A1Y1MZN7 A0A1Y1MWE3 A0A1Y1MUU2 A0A1Y1MV75 A0A1Y1MUT0 A0A026WS19 A0A1B6DGI9 E0V9G5 D1ZZH5 A0A3L8DXG6 A0A2R7VPE6 A0A1B6LAP2 E9ISS0 A0A2J7PYW9 A0A2J7PYZ3 A0A088AGX0 A0A151IH48 A0A2J7PYZ1 A0A158NPE3 A0A0L7QK28 A0A195F4F0 A0A0A9ZII9 A0A224XTW4 A0A195E5B4 F4WH76 A0A2A3ESC0 A0A195B9D0 A0A023F394 E2ADU8 A0A154P6Q6 A0A0V0GCB0 A0A151X644 A0A0N0BFU8 A0A226E6R1 A0A1B6JM49 A0A0C9PJQ9 A0A1L8DHN4 A0A444TE91 A0A443QII4 A0A2D0PLM0 A0A131XG30 A0A131YQM8 A0A443SJD6 A0A2D0PJ21 A0A2D0PJ18 A0A224YUZ2 A0A2H8THC0 G1KSQ3 A0A1Q3F594 A0A1Q3F4F3 A0A0P4WE93 A0A3S2M950 A0A3P9H587 A0A3Q2XFX0 A0A2D0PLM6 A0A402EK00 B0R049 A0A3Q1FZ19 J9LW24 A0A315W4Z2 A0A1E1XDW9 A0A3Q3LQW8 A0A3B4WP72 A0A1A8HIU3 A0A1A8P5W2 A0A1A7ZBK8 A0A3Q3G304 A0A151P519 A0A1A7XWH7 V8P2H7 A0A2G8K6E2
S4P253 A0A212FDX3 A0A2A4IYZ3 A0A194R7J7 A0A1W4WP73 A0A1W4WP55 A0A1Y1MZN7 A0A1Y1MWE3 A0A1Y1MUU2 A0A1Y1MV75 A0A1Y1MUT0 A0A026WS19 A0A1B6DGI9 E0V9G5 D1ZZH5 A0A3L8DXG6 A0A2R7VPE6 A0A1B6LAP2 E9ISS0 A0A2J7PYW9 A0A2J7PYZ3 A0A088AGX0 A0A151IH48 A0A2J7PYZ1 A0A158NPE3 A0A0L7QK28 A0A195F4F0 A0A0A9ZII9 A0A224XTW4 A0A195E5B4 F4WH76 A0A2A3ESC0 A0A195B9D0 A0A023F394 E2ADU8 A0A154P6Q6 A0A0V0GCB0 A0A151X644 A0A0N0BFU8 A0A226E6R1 A0A1B6JM49 A0A0C9PJQ9 A0A1L8DHN4 A0A444TE91 A0A443QII4 A0A2D0PLM0 A0A131XG30 A0A131YQM8 A0A443SJD6 A0A2D0PJ21 A0A2D0PJ18 A0A224YUZ2 A0A2H8THC0 G1KSQ3 A0A1Q3F594 A0A1Q3F4F3 A0A0P4WE93 A0A3S2M950 A0A3P9H587 A0A3Q2XFX0 A0A2D0PLM6 A0A402EK00 B0R049 A0A3Q1FZ19 J9LW24 A0A315W4Z2 A0A1E1XDW9 A0A3Q3LQW8 A0A3B4WP72 A0A1A8HIU3 A0A1A8P5W2 A0A1A7ZBK8 A0A3Q3G304 A0A151P519 A0A1A7XWH7 V8P2H7 A0A2G8K6E2
PDB
4AWL
E-value=2.66296e-16,
Score=207
Ontologies
GO
PANTHER
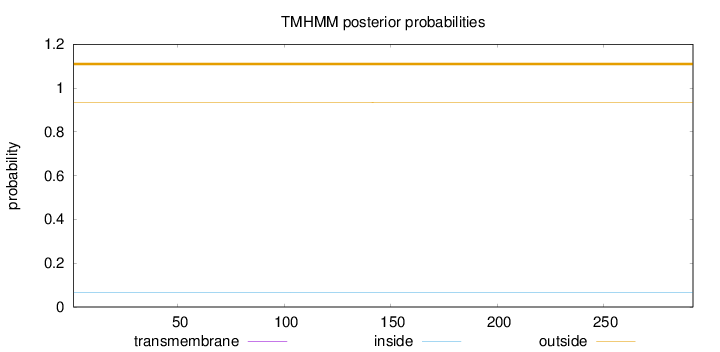
Topology
Length:
292
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00226
Exp number, first 60 AAs:
0.00055
Total prob of N-in:
0.06689
outside
1 - 292
Population Genetic Test Statistics
Pi
168.564194
Theta
161.124605
Tajima's D
0.158545
CLR
0.90205
CSRT
0.410829458527074
Interpretation
Uncertain
