Pre Gene Modal
BGIBMGA009240
Annotation
PREDICTED:_N-alpha-acetyltransferase_15?_NatA_auxiliary_subunit_[Papilio_xuthus]
Full name
Glycerol-3-phosphate dehydrogenase [NAD(+)]
Location in the cell
Cytoplasmic Reliability : 1.761 Mitochondrial Reliability : 1.203 Nuclear Reliability : 1.495
Sequence
CDS
ATGCCTCCAAGCAATCCTTTACCACCTAAAGAGAATGCTCTTTTTAAAAGAATTTTGCGGTGCTACGAACATAAACAATACAAAAATGGGTTAAAATTCGCCAAACAGATATTATCCAACCCAAAGTTCGCGGAGCATGGAGAAACCTTAGCCATGAAAGGTCTAACATTAAATTGCTTAGGTCGCAAGGATGAGGCATATGACTATGTACGCAGAGGTCTACGTAATGATCTGAAGTCCCCAGTATGTTGGCATGTGTATGGGCTGTTGCAGCGCTCTGACAAGAAATACGATGAGGCAATAAAATGTTACCGTAATGCACTAAAATGGGAAAAGGAGAATATTCAAATTCTCAGAGATTTGTCACTATTACAAATTCAGATGAGAGATTTAGAGGGTTATAAAGATACCAGATATCAGCTTTTCATGCTGAGGCCTACACAAAGAGCATCATGGATAGGATTTGCTATGAGCTACCATCTCCTCGGTGATTATGAAATGGCAAACAGTATCCTAGATGCATTCCGCACTCATCAGATGAAAGGTCCATATGATTATGAACACTCAGAATTGCTATTATACCAGAATATGGTACTGGCCGAATCAGGACAATATGAGAGAGCCTTGCAGCATTTACAGAAGTTTGAAAGTCAAATTCTGGATAAATTATCTGTGAAAGAAACTTCTGGGGAATATTACTTAAAATTGAACAGATTCAAGGAAGCAGAAGGAATTTACGAAGACTTGTTAAAGAGAAATCCCGAAAATGTTTTGTACTACAGTAAGTTAATAGAAGCGAAACAACTCATTAAAGCTGAAGACAAAGTGGCCTTTTTTGATGAATACAAGAAGCAGTATCCAAAGGCAATAGCACCGCGACGTCTGCAATTGACCGAGGCACAGGCGCAGTCCTCATTTGAAAACCTCGTTGACGAGTACCTCAGACATGGACTGTACAAAGGAATTCCTCCACTTTTTGTAGATTTGAGATCTTTATACGCAGATCCAATGAAAGCGGAAACGATAGAGAAATTAATAATACAGTACATAGATAATTTAGCTAAAGATTGTACATTTGGACCAGACCCGAGTGAAGTGAAGCAGCCCGCAAGTGCGCTTTTGTGGGCGTACTATTTCGCGGCCCAACACTTCGATCACAAGAAGGATACGGATCGAGCGTTGCATTACATAGACGCCGCCATTGAACACACCCCTACATTGATAGAATTGTTTATTGTTAAGGGAAGAATATACAAGCATGCCGGCGACCCAATATCCGCGTACCAGTGCTTAGAGGAAGCTCAAGCGATGGACACGGCGGATCGATACGTCAACTGTAAATGCGCGCGGTACATGCTCAGGGCGGGCCACATCAAACGAGCCGAGGAGATGTGCGCCAAGTTCACGAGGGAAGGTGTACCGGCAACGGAAAACTTAAATGAAATGCAATGTATGTGGTTCCAAACGGAAGCTGCTGCTTCATACCAAAGACTTCAACAATGGGGAGAGGCTCTGAAAAAAGCACACGAAGTCGACAGACACTTCTCGGAGATAATGGAGGACCAGTTCGACTTCCACTCGTACTGCATGCGCAAGATGACGCTCCGCTCGTACGTGGGGCTGCTGCGGCTGGAGGACGTGCTGCGCGCGCACCCCTTCTACTTCCGCTGCGCGCGCGTCGCCGTGCGCGTGTACCTGCGCCTGCACGCGCGCCCGCGCTCCGACGCGCCGCAGACCGCCGAGCCCGACACAGGTATGTCGCCATCGTACATACATCATTACTAG
Protein
MPPSNPLPPKENALFKRILRCYEHKQYKNGLKFAKQILSNPKFAEHGETLAMKGLTLNCLGRKDEAYDYVRRGLRNDLKSPVCWHVYGLLQRSDKKYDEAIKCYRNALKWEKENIQILRDLSLLQIQMRDLEGYKDTRYQLFMLRPTQRASWIGFAMSYHLLGDYEMANSILDAFRTHQMKGPYDYEHSELLLYQNMVLAESGQYERALQHLQKFESQILDKLSVKETSGEYYLKLNRFKEAEGIYEDLLKRNPENVLYYSKLIEAKQLIKAEDKVAFFDEYKKQYPKAIAPRRLQLTEAQAQSSFENLVDEYLRHGLYKGIPPLFVDLRSLYADPMKAETIEKLIIQYIDNLAKDCTFGPDPSEVKQPASALLWAYYFAAQHFDHKKDTDRALHYIDAAIEHTPTLIELFIVKGRIYKHAGDPISAYQCLEEAQAMDTADRYVNCKCARYMLRAGHIKRAEEMCAKFTREGVPATENLNEMQCMWFQTEAAASYQRLQQWGEALKKAHEVDRHFSEIMEDQFDFHSYCMRKMTLRSYVGLLRLEDVLRAHPFYFRCARVAVRVYLRLHARPRSDAPQTAEPDTGMSPSYIHHY
Summary
Catalytic Activity
NAD(+) + sn-glycerol 3-phosphate = dihydroxyacetone phosphate + H(+) + NADH
Similarity
Belongs to the NAD-dependent glycerol-3-phosphate dehydrogenase family.
Uniprot
H9JI90
A0A2W1BSB1
A0A2H1VJ79
A0A194PXZ9
A0A212FDY8
A0A067R5Y7
+ More
A0A1B6HMF7 A0A1W4XAU5 A0A2A3EQW5 A0A1W4XBZ6 A0A087ZRP6 D2A0B9 A0A0C9R5T2 A0A232F5T2 K7J6S9 A0A0N0BI27 A0A195CGQ8 A0A195BGR2 A0A158N9D6 A0A151XIC8 A0A336KKR7 A0A2H1VJ75 A0A1B6C9Q1 A0A336M640 E2ANZ3 A0A1Y1N7A8 A0A0J7L4Z6 A0A3Q0ISG2 F4WJY7 A0A146KLI1 E9IP68 A0A0A9ZBP9 A0A1Y1N751 A0A146LKY9 A0A0K8SFC9 E2BPZ9 A0A3L8DR34 A0A0T6B9Q7 E0VDK3 A0A1Y1N767 A0A0L7R5S2 A0A182HEW7 Q177P1 V5IK41 A0A026VVM1 A0A1E1X2E2 L7MED8 A0A087U3V4 A0A182NG76 A0A131Z4E0 A0A195F9H2 U5EYF6 A0A224YRD0 A0A2H8TV72 A0A131XHZ7 A0A0L0C6F5 A0A1I8MMP8 A0A2S2R005 A0A195E5W2 A0A1B0G1J1 A0A1A9UXZ8 A0A1A9Z1V2 B4NPI8 A0A1A9XRZ5 A0A1B0BK08 A0A182QY12 A0A2M4A9P2 A0A1I8NSN7 W5JUB5 A0A2M3ZGD5 A0A2M3Z1S0 A0A182FPY5 A0A2M4BDP0 Q7QKE3 A0A182HS16 A0A0M4F9R1 A0A1A9WXX6 X1WIP8 A0A182LQD4 A0A1L8DWR9 A0A182UP71 A0A182SFX9 A0A182KDU1 A0A1L8DWX1 A0A1J1J5C4 A0A293N4L4 A0A182W2K4 A0A182R7M4 E9FTX7 A0A182XVC6 B4MA47 A0A0K8WFG9 B3NVN4 A0A0A1X689 A0A182PEH1 Q9VWI2 A0A034VJH4 A0A182J3Q8 B4PZI3 A0A2R5LMW1
A0A1B6HMF7 A0A1W4XAU5 A0A2A3EQW5 A0A1W4XBZ6 A0A087ZRP6 D2A0B9 A0A0C9R5T2 A0A232F5T2 K7J6S9 A0A0N0BI27 A0A195CGQ8 A0A195BGR2 A0A158N9D6 A0A151XIC8 A0A336KKR7 A0A2H1VJ75 A0A1B6C9Q1 A0A336M640 E2ANZ3 A0A1Y1N7A8 A0A0J7L4Z6 A0A3Q0ISG2 F4WJY7 A0A146KLI1 E9IP68 A0A0A9ZBP9 A0A1Y1N751 A0A146LKY9 A0A0K8SFC9 E2BPZ9 A0A3L8DR34 A0A0T6B9Q7 E0VDK3 A0A1Y1N767 A0A0L7R5S2 A0A182HEW7 Q177P1 V5IK41 A0A026VVM1 A0A1E1X2E2 L7MED8 A0A087U3V4 A0A182NG76 A0A131Z4E0 A0A195F9H2 U5EYF6 A0A224YRD0 A0A2H8TV72 A0A131XHZ7 A0A0L0C6F5 A0A1I8MMP8 A0A2S2R005 A0A195E5W2 A0A1B0G1J1 A0A1A9UXZ8 A0A1A9Z1V2 B4NPI8 A0A1A9XRZ5 A0A1B0BK08 A0A182QY12 A0A2M4A9P2 A0A1I8NSN7 W5JUB5 A0A2M3ZGD5 A0A2M3Z1S0 A0A182FPY5 A0A2M4BDP0 Q7QKE3 A0A182HS16 A0A0M4F9R1 A0A1A9WXX6 X1WIP8 A0A182LQD4 A0A1L8DWR9 A0A182UP71 A0A182SFX9 A0A182KDU1 A0A1L8DWX1 A0A1J1J5C4 A0A293N4L4 A0A182W2K4 A0A182R7M4 E9FTX7 A0A182XVC6 B4MA47 A0A0K8WFG9 B3NVN4 A0A0A1X689 A0A182PEH1 Q9VWI2 A0A034VJH4 A0A182J3Q8 B4PZI3 A0A2R5LMW1
EC Number
1.1.1.8
Pubmed
19121390
28756777
26354079
22118469
24845553
18362917
+ More
19820115 28648823 20075255 21347285 20798317 28004739 21719571 26823975 21282665 25401762 30249741 20566863 26483478 17510324 25765539 24508170 28503490 25576852 26830274 28797301 28049606 26108605 25315136 17994087 20920257 23761445 12364791 14747013 17210077 20966253 21292972 25244985 18057021 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 17550304
19820115 28648823 20075255 21347285 20798317 28004739 21719571 26823975 21282665 25401762 30249741 20566863 26483478 17510324 25765539 24508170 28503490 25576852 26830274 28797301 28049606 26108605 25315136 17994087 20920257 23761445 12364791 14747013 17210077 20966253 21292972 25244985 18057021 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 17550304
EMBL
BABH01022256
BABH01022257
KZ149916
PZC77948.1
ODYU01002868
SOQ40883.1
+ More
KQ459585 KPI98216.1 AGBW02008992 OWR51952.1 KK852672 KDR18769.1 GECU01031862 JAS75844.1 KZ288202 PBC33529.1 KQ971338 EFA02495.1 GBYB01003290 GBYB01003291 JAG73057.1 JAG73058.1 NNAY01000856 OXU26184.1 AAZX01000215 KQ435737 KOX76948.1 KQ977791 KYM99915.1 KQ976476 KYM83848.1 ADTU01009482 ADTU01009483 ADTU01009484 KQ982093 KYQ60071.1 UFQS01000513 UFQT01000513 SSX04518.1 SSX24881.1 ODYU01002869 SOQ40885.1 GEDC01027081 JAS10217.1 UFQS01000608 UFQT01000608 SSX05324.1 SSX25685.1 GL441439 EFN64825.1 GEZM01010886 JAV93751.1 LBMM01000733 KMQ97613.1 GL888187 EGI65509.1 GDHC01021208 JAP97420.1 GL764458 EFZ17636.1 GBHO01001736 GBHO01001735 JAG41868.1 JAG41869.1 GEZM01010884 JAV93754.1 GDHC01010001 JAQ08628.1 GBRD01013982 JAG51844.1 GL449694 EFN82187.1 QOIP01000005 RLU22258.1 LJIG01002850 KRT84090.1 DS235078 EEB11459.1 GEZM01010885 JAV93752.1 KQ414648 KOC66230.1 JXUM01132778 KQ567891 KXJ69234.1 CH477373 EAT42377.1 GANP01000347 JAB84121.1 KK108118 EZA46879.1 GFAC01005765 JAT93423.1 GACK01002669 JAA62365.1 KK118034 KFM72043.1 GEDV01003566 JAP84991.1 KQ981727 KYN36699.1 GANO01002031 JAB57840.1 GFPF01005677 MAA16823.1 GFXV01006312 MBW18117.1 GEFH01001828 JAP66753.1 JRES01000940 KNC27009.1 GGMS01014138 MBY83341.1 KQ979592 KYN20585.1 CCAG010002137 CH964291 EDW86428.1 JXJN01015768 AXCN02001630 GGFK01004203 MBW37524.1 ADMH02000471 ETN66349.1 GGFM01006794 MBW27545.1 GGFM01001702 MBW22453.1 GGFJ01001787 MBW50928.1 AAAB01008799 EAA03779.3 APCN01000232 CP012528 ALC49337.1 ABLF02040094 ABLF02040101 ABLF02040106 ABLF02040107 GFDF01003184 JAV10900.1 GFDF01003185 JAV10899.1 CVRI01000073 CRL07613.1 GFWV01022622 MAA47349.1 GL732524 EFX89563.1 CH940655 EDW66106.1 KRF82563.1 GDHF01033695 GDHF01031825 GDHF01027382 GDHF01015353 GDHF01004443 GDHF01002482 GDHF01002480 GDHF01000171 JAI18619.1 JAI20489.1 JAI24932.1 JAI36961.1 JAI47871.1 JAI49832.1 JAI49834.1 JAI52143.1 CH954180 EDV46699.1 GBXI01007901 JAD06391.1 AE014298 BT014631 AAF48957.1 AAT27255.1 AHN59909.1 GAKP01016707 GAKP01016705 JAC42247.1 CM000162 EDX02139.1 GGLE01006747 MBY10873.1
KQ459585 KPI98216.1 AGBW02008992 OWR51952.1 KK852672 KDR18769.1 GECU01031862 JAS75844.1 KZ288202 PBC33529.1 KQ971338 EFA02495.1 GBYB01003290 GBYB01003291 JAG73057.1 JAG73058.1 NNAY01000856 OXU26184.1 AAZX01000215 KQ435737 KOX76948.1 KQ977791 KYM99915.1 KQ976476 KYM83848.1 ADTU01009482 ADTU01009483 ADTU01009484 KQ982093 KYQ60071.1 UFQS01000513 UFQT01000513 SSX04518.1 SSX24881.1 ODYU01002869 SOQ40885.1 GEDC01027081 JAS10217.1 UFQS01000608 UFQT01000608 SSX05324.1 SSX25685.1 GL441439 EFN64825.1 GEZM01010886 JAV93751.1 LBMM01000733 KMQ97613.1 GL888187 EGI65509.1 GDHC01021208 JAP97420.1 GL764458 EFZ17636.1 GBHO01001736 GBHO01001735 JAG41868.1 JAG41869.1 GEZM01010884 JAV93754.1 GDHC01010001 JAQ08628.1 GBRD01013982 JAG51844.1 GL449694 EFN82187.1 QOIP01000005 RLU22258.1 LJIG01002850 KRT84090.1 DS235078 EEB11459.1 GEZM01010885 JAV93752.1 KQ414648 KOC66230.1 JXUM01132778 KQ567891 KXJ69234.1 CH477373 EAT42377.1 GANP01000347 JAB84121.1 KK108118 EZA46879.1 GFAC01005765 JAT93423.1 GACK01002669 JAA62365.1 KK118034 KFM72043.1 GEDV01003566 JAP84991.1 KQ981727 KYN36699.1 GANO01002031 JAB57840.1 GFPF01005677 MAA16823.1 GFXV01006312 MBW18117.1 GEFH01001828 JAP66753.1 JRES01000940 KNC27009.1 GGMS01014138 MBY83341.1 KQ979592 KYN20585.1 CCAG010002137 CH964291 EDW86428.1 JXJN01015768 AXCN02001630 GGFK01004203 MBW37524.1 ADMH02000471 ETN66349.1 GGFM01006794 MBW27545.1 GGFM01001702 MBW22453.1 GGFJ01001787 MBW50928.1 AAAB01008799 EAA03779.3 APCN01000232 CP012528 ALC49337.1 ABLF02040094 ABLF02040101 ABLF02040106 ABLF02040107 GFDF01003184 JAV10900.1 GFDF01003185 JAV10899.1 CVRI01000073 CRL07613.1 GFWV01022622 MAA47349.1 GL732524 EFX89563.1 CH940655 EDW66106.1 KRF82563.1 GDHF01033695 GDHF01031825 GDHF01027382 GDHF01015353 GDHF01004443 GDHF01002482 GDHF01002480 GDHF01000171 JAI18619.1 JAI20489.1 JAI24932.1 JAI36961.1 JAI47871.1 JAI49832.1 JAI49834.1 JAI52143.1 CH954180 EDV46699.1 GBXI01007901 JAD06391.1 AE014298 BT014631 AAF48957.1 AAT27255.1 AHN59909.1 GAKP01016707 GAKP01016705 JAC42247.1 CM000162 EDX02139.1 GGLE01006747 MBY10873.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000027135
UP000192223
UP000242457
+ More
UP000005203 UP000007266 UP000215335 UP000002358 UP000053105 UP000078542 UP000078540 UP000005205 UP000075809 UP000000311 UP000036403 UP000079169 UP000007755 UP000008237 UP000279307 UP000009046 UP000053825 UP000069940 UP000249989 UP000008820 UP000053097 UP000054359 UP000075884 UP000078541 UP000037069 UP000095301 UP000078492 UP000092444 UP000078200 UP000092445 UP000007798 UP000092443 UP000092460 UP000075886 UP000095300 UP000000673 UP000069272 UP000007062 UP000075840 UP000092553 UP000091820 UP000007819 UP000075882 UP000075903 UP000075901 UP000075881 UP000183832 UP000075920 UP000075900 UP000000305 UP000076408 UP000008792 UP000008711 UP000075885 UP000000803 UP000075880 UP000002282
UP000005203 UP000007266 UP000215335 UP000002358 UP000053105 UP000078542 UP000078540 UP000005205 UP000075809 UP000000311 UP000036403 UP000079169 UP000007755 UP000008237 UP000279307 UP000009046 UP000053825 UP000069940 UP000249989 UP000008820 UP000053097 UP000054359 UP000075884 UP000078541 UP000037069 UP000095301 UP000078492 UP000092444 UP000078200 UP000092445 UP000007798 UP000092443 UP000092460 UP000075886 UP000095300 UP000000673 UP000069272 UP000007062 UP000075840 UP000092553 UP000091820 UP000007819 UP000075882 UP000075903 UP000075901 UP000075881 UP000183832 UP000075920 UP000075900 UP000000305 UP000076408 UP000008792 UP000008711 UP000075885 UP000000803 UP000075880 UP000002282
PRIDE
Pfam
Interpro
IPR021183
NatA_aux_su
+ More
IPR013026 TPR-contain_dom
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR017853 Glycoside_hydrolase_SF
IPR011496 Beta-N-acetylglucosaminidase
IPR036291 NAD(P)-bd_dom_sf
IPR008927 6-PGluconate_DH-like_C_sf
IPR006168 G3P_DH_NAD-dep
IPR013328 6PGD_dom2
IPR011128 G3P_DH_NAD-dep_N
IPR006109 G3P_DH_NAD-dep_C
IPR017751 G3P_DH_NAD-dep_euk
IPR013026 TPR-contain_dom
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR017853 Glycoside_hydrolase_SF
IPR011496 Beta-N-acetylglucosaminidase
IPR036291 NAD(P)-bd_dom_sf
IPR008927 6-PGluconate_DH-like_C_sf
IPR006168 G3P_DH_NAD-dep
IPR013328 6PGD_dom2
IPR011128 G3P_DH_NAD-dep_N
IPR006109 G3P_DH_NAD-dep_C
IPR017751 G3P_DH_NAD-dep_euk
Gene 3D
ProteinModelPortal
H9JI90
A0A2W1BSB1
A0A2H1VJ79
A0A194PXZ9
A0A212FDY8
A0A067R5Y7
+ More
A0A1B6HMF7 A0A1W4XAU5 A0A2A3EQW5 A0A1W4XBZ6 A0A087ZRP6 D2A0B9 A0A0C9R5T2 A0A232F5T2 K7J6S9 A0A0N0BI27 A0A195CGQ8 A0A195BGR2 A0A158N9D6 A0A151XIC8 A0A336KKR7 A0A2H1VJ75 A0A1B6C9Q1 A0A336M640 E2ANZ3 A0A1Y1N7A8 A0A0J7L4Z6 A0A3Q0ISG2 F4WJY7 A0A146KLI1 E9IP68 A0A0A9ZBP9 A0A1Y1N751 A0A146LKY9 A0A0K8SFC9 E2BPZ9 A0A3L8DR34 A0A0T6B9Q7 E0VDK3 A0A1Y1N767 A0A0L7R5S2 A0A182HEW7 Q177P1 V5IK41 A0A026VVM1 A0A1E1X2E2 L7MED8 A0A087U3V4 A0A182NG76 A0A131Z4E0 A0A195F9H2 U5EYF6 A0A224YRD0 A0A2H8TV72 A0A131XHZ7 A0A0L0C6F5 A0A1I8MMP8 A0A2S2R005 A0A195E5W2 A0A1B0G1J1 A0A1A9UXZ8 A0A1A9Z1V2 B4NPI8 A0A1A9XRZ5 A0A1B0BK08 A0A182QY12 A0A2M4A9P2 A0A1I8NSN7 W5JUB5 A0A2M3ZGD5 A0A2M3Z1S0 A0A182FPY5 A0A2M4BDP0 Q7QKE3 A0A182HS16 A0A0M4F9R1 A0A1A9WXX6 X1WIP8 A0A182LQD4 A0A1L8DWR9 A0A182UP71 A0A182SFX9 A0A182KDU1 A0A1L8DWX1 A0A1J1J5C4 A0A293N4L4 A0A182W2K4 A0A182R7M4 E9FTX7 A0A182XVC6 B4MA47 A0A0K8WFG9 B3NVN4 A0A0A1X689 A0A182PEH1 Q9VWI2 A0A034VJH4 A0A182J3Q8 B4PZI3 A0A2R5LMW1
A0A1B6HMF7 A0A1W4XAU5 A0A2A3EQW5 A0A1W4XBZ6 A0A087ZRP6 D2A0B9 A0A0C9R5T2 A0A232F5T2 K7J6S9 A0A0N0BI27 A0A195CGQ8 A0A195BGR2 A0A158N9D6 A0A151XIC8 A0A336KKR7 A0A2H1VJ75 A0A1B6C9Q1 A0A336M640 E2ANZ3 A0A1Y1N7A8 A0A0J7L4Z6 A0A3Q0ISG2 F4WJY7 A0A146KLI1 E9IP68 A0A0A9ZBP9 A0A1Y1N751 A0A146LKY9 A0A0K8SFC9 E2BPZ9 A0A3L8DR34 A0A0T6B9Q7 E0VDK3 A0A1Y1N767 A0A0L7R5S2 A0A182HEW7 Q177P1 V5IK41 A0A026VVM1 A0A1E1X2E2 L7MED8 A0A087U3V4 A0A182NG76 A0A131Z4E0 A0A195F9H2 U5EYF6 A0A224YRD0 A0A2H8TV72 A0A131XHZ7 A0A0L0C6F5 A0A1I8MMP8 A0A2S2R005 A0A195E5W2 A0A1B0G1J1 A0A1A9UXZ8 A0A1A9Z1V2 B4NPI8 A0A1A9XRZ5 A0A1B0BK08 A0A182QY12 A0A2M4A9P2 A0A1I8NSN7 W5JUB5 A0A2M3ZGD5 A0A2M3Z1S0 A0A182FPY5 A0A2M4BDP0 Q7QKE3 A0A182HS16 A0A0M4F9R1 A0A1A9WXX6 X1WIP8 A0A182LQD4 A0A1L8DWR9 A0A182UP71 A0A182SFX9 A0A182KDU1 A0A1L8DWX1 A0A1J1J5C4 A0A293N4L4 A0A182W2K4 A0A182R7M4 E9FTX7 A0A182XVC6 B4MA47 A0A0K8WFG9 B3NVN4 A0A0A1X689 A0A182PEH1 Q9VWI2 A0A034VJH4 A0A182J3Q8 B4PZI3 A0A2R5LMW1
PDB
6C9M
E-value=0,
Score=1863
Ontologies
GO
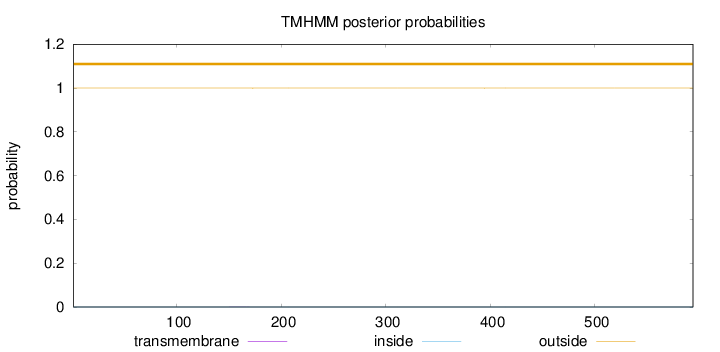
Topology
Length:
594
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00500999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00018
outside
1 - 594
Population Genetic Test Statistics
Pi
221.294451
Theta
153.707974
Tajima's D
0.419616
CLR
0.823035
CSRT
0.492575371231438
Interpretation
Uncertain
