Pre Gene Modal
BGIBMGA009241
Annotation
PREDICTED:_N-alpha-acetyltransferase_15?_NatA_auxiliary_subunit_[Papilio_xuthus]
Full name
Glycerol-3-phosphate dehydrogenase [NAD(+)]
Location in the cell
Cytoplasmic Reliability : 2.233
Sequence
CDS
ATGGGAACTAAAAAAAAAAACCTAGCGCCTTCAGAATTAAAAAAGCTTAGAAATAAACAAAGGAAAGCCCAGCGTAAAGCAGAGCAAGAAAGCGCTCTGGCGGCACAAGTACAGGTAAAACGGGAGCAGCACCATAAAGCGCGGCAACAGCAGGAACAAGGCGACCCCGAAGCGCCGCAACTCGACGAACTGATACCCGAGAAACTAGCGAGAGTAGAAGATCCATTAGAACAAGCATTGAAATTCTTGCAGCCTCTGAGGATACTCGCGGCGGACAGGATAGAGACACACCTAATGGCCTTCGAAATTTACTTTAGAAAAGAAAAGCCCCTACTGATGCTGCAGTGTATAAAGAGAGCGTGGAGATTGGACAGCGATAATCCTGAGCTGGTGGACTGTCTGGTTCGTTTCCAACGGTGGCTGGAGGAATCGCTTTCCGGGCTGCATCCGAACGTTGCGGCCGTTCTTAAGACTGAGACCCAACCGATGGTGTTGGGACGCAGCGCGCTGCAGATCGCGGAGCAGAGCGTGTCGGGCGCGGCGCGGCACGCGGCGGCGCTGTGGGCGGCGCGCGCGCTGCGGCGGCTCGTGCCCGACCGCACGGAGCAGGCGCTGCGCCTCGTCACCTCGCTCGACTACCCGGACGTCACTATACAGAGTTGCGCAGACGTTTTAGAGAGCCTCAGAGAAGGCGATTTCGGTGAATGCGAGGCGGTGATTGAGCGCTACGTGGAGGCGTGCAGCGCGCGGTTCCCGTACGCGATCGCCTTCAAGCCCGCGCCCGCGCCCGAAGCGAGCGAGAACCATTCGGAAGAGACTACACCGCTACAGCCTAAAGAAATTACTGCCAACAACTAA
Protein
MGTKKKNLAPSELKKLRNKQRKAQRKAEQESALAAQVQVKREQHHKARQQQEQGDPEAPQLDELIPEKLARVEDPLEQALKFLQPLRILAADRIETHLMAFEIYFRKEKPLLMLQCIKRAWRLDSDNPELVDCLVRFQRWLEESLSGLHPNVAAVLKTETQPMVLGRSALQIAEQSVSGAARHAAALWAARALRRLVPDRTEQALRLVTSLDYPDVTIQSCADVLESLREGDFGECEAVIERYVEACSARFPYAIAFKPAPAPEASENHSEETTPLQPKEITANN
Summary
Catalytic Activity
NAD(+) + sn-glycerol 3-phosphate = dihydroxyacetone phosphate + H(+) + NADH
Similarity
Belongs to the NAD-dependent glycerol-3-phosphate dehydrogenase family.
Uniprot
H9JI91
A0A2W1BZ33
A0A2H1W3K0
A0A437BS50
D2A0B9
F4WJY5
+ More
E9IP70 A0A0L7R5V7 A0A232F6C0 E2ANZ6 N6UMP0 E2BPZ7 A0A2J7PPV0 A0A0J7L4R9 N6TQQ6 A0A0N0U6I2 A0A087ZRP7 A0A1B0FY46 A0A158N9D6 A0A026VVM1 A0A3L8DR34 A0A195BGR2 A0A195CGQ8 A0A151XIC8 A0A195E5W2 A0A195F9H2 A0A0C9R5T2 A0A1Y1N751 A0A1W4XAU5 A0A2A3EQW5 A0A1L8DWR9 A0A1L8DWX1 A0A1B0CM67 V5GPW5 W8BNC4 A0A0L0C6F5 A0A0A1X689 A0A1A9WXX6 A0A336KKR7 T1P942 A0A1I8MMP8 A0A1A9XRZ5 A0A1B0BK08 U5EYF6 A0A1B0G1J1 A0A034VJH4 A0A1A9Z1V2 E0VDK3 A0A1I8NSN7 A0A1A9UXZ8 T1INF4 A0A0K8WFG9 B3MPT7 A0A2S2NLB3 B4PZI3 Q9VWI2 B3NVN4 A0A1W4VTE2 B4NPI8 A0A2S2R005 X1WIP8 A0A1B6MQ90 B4L6X3 Q29IV7 B4GW65 A0A2H8TV72 A0A182HEW7 A0A3B0KTD8 A0A3B0K712 A0A023F378 A0A087U3V4 A0A2R7WIZ7 A0A0M4F9R1 A0A336M640 A0A1Q3F3K0 A0A182QY12 A0A1S3JLG3 A0A1B6F8C2 Q177P1 A0A069DZM3 A0A1B6EDW5 A0A224XED0 A0A1W4XBZ6 B4MA47 A0A1J1J5C4 B7P5N3 A0A0P4VJF5 B4JX27 A0A1Y1N7A8 A0A0P6FVD5 A0A182NG76 A0A0N8CFZ0 A0A0P4YQ39 A0A0P5YCM2 A0A0P5QYP9 A0A0P6HXS5 V5IK41
E9IP70 A0A0L7R5V7 A0A232F6C0 E2ANZ6 N6UMP0 E2BPZ7 A0A2J7PPV0 A0A0J7L4R9 N6TQQ6 A0A0N0U6I2 A0A087ZRP7 A0A1B0FY46 A0A158N9D6 A0A026VVM1 A0A3L8DR34 A0A195BGR2 A0A195CGQ8 A0A151XIC8 A0A195E5W2 A0A195F9H2 A0A0C9R5T2 A0A1Y1N751 A0A1W4XAU5 A0A2A3EQW5 A0A1L8DWR9 A0A1L8DWX1 A0A1B0CM67 V5GPW5 W8BNC4 A0A0L0C6F5 A0A0A1X689 A0A1A9WXX6 A0A336KKR7 T1P942 A0A1I8MMP8 A0A1A9XRZ5 A0A1B0BK08 U5EYF6 A0A1B0G1J1 A0A034VJH4 A0A1A9Z1V2 E0VDK3 A0A1I8NSN7 A0A1A9UXZ8 T1INF4 A0A0K8WFG9 B3MPT7 A0A2S2NLB3 B4PZI3 Q9VWI2 B3NVN4 A0A1W4VTE2 B4NPI8 A0A2S2R005 X1WIP8 A0A1B6MQ90 B4L6X3 Q29IV7 B4GW65 A0A2H8TV72 A0A182HEW7 A0A3B0KTD8 A0A3B0K712 A0A023F378 A0A087U3V4 A0A2R7WIZ7 A0A0M4F9R1 A0A336M640 A0A1Q3F3K0 A0A182QY12 A0A1S3JLG3 A0A1B6F8C2 Q177P1 A0A069DZM3 A0A1B6EDW5 A0A224XED0 A0A1W4XBZ6 B4MA47 A0A1J1J5C4 B7P5N3 A0A0P4VJF5 B4JX27 A0A1Y1N7A8 A0A0P6FVD5 A0A182NG76 A0A0N8CFZ0 A0A0P4YQ39 A0A0P5YCM2 A0A0P5QYP9 A0A0P6HXS5 V5IK41
EC Number
1.1.1.8
Pubmed
19121390
28756777
18362917
19820115
21719571
21282665
+ More
28648823 20798317 23537049 21347285 24508170 30249741 28004739 24495485 26108605 25830018 25315136 25348373 20566863 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 23185243 26483478 25474469 17510324 26334808 27129103 25765539
28648823 20798317 23537049 21347285 24508170 30249741 28004739 24495485 26108605 25830018 25315136 25348373 20566863 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 23185243 26483478 25474469 17510324 26334808 27129103 25765539
EMBL
BABH01022251
KZ149916
PZC77946.1
ODYU01005810
SOQ47074.1
RSAL01000016
+ More
RVE53059.1 KQ971338 EFA02495.1 GL888187 EGI65507.1 GL764458 EFZ17627.1 KQ414648 KOC66228.1 NNAY01000856 OXU26185.1 GL441439 EFN64828.1 APGK01008041 KB737564 KB632337 ENN83000.1 ERL92856.1 GL449694 EFN82185.1 NEVH01022644 PNF18368.1 LBMM01000733 KMQ97611.1 APGK01009386 KB737932 ENN82834.1 KQ435737 KOX76951.1 AJVK01005172 AJVK01005173 AJVK01005174 ADTU01009482 ADTU01009483 ADTU01009484 KK108118 EZA46879.1 QOIP01000005 RLU22258.1 KQ976476 KYM83848.1 KQ977791 KYM99915.1 KQ982093 KYQ60071.1 KQ979592 KYN20585.1 KQ981727 KYN36699.1 GBYB01003290 GBYB01003291 JAG73057.1 JAG73058.1 GEZM01010884 JAV93754.1 KZ288202 PBC33529.1 GFDF01003184 JAV10900.1 GFDF01003185 JAV10899.1 AJWK01018284 AJWK01018285 AJWK01018286 AJWK01018287 GALX01002292 JAB66174.1 GAMC01006239 GAMC01006238 JAC00318.1 JRES01000940 KNC27009.1 GBXI01007901 JAD06391.1 UFQS01000513 UFQT01000513 SSX04518.1 SSX24881.1 KA645227 AFP59856.1 JXJN01015768 GANO01002031 JAB57840.1 CCAG010002137 GAKP01016707 GAKP01016705 JAC42247.1 DS235078 EEB11459.1 JH431165 GDHF01033695 GDHF01031825 GDHF01027382 GDHF01015353 GDHF01004443 GDHF01002482 GDHF01002480 GDHF01000171 JAI18619.1 JAI20489.1 JAI24932.1 JAI36961.1 JAI47871.1 JAI49832.1 JAI49834.1 JAI52143.1 CH902621 EDV44699.1 GGMR01005361 MBY17980.1 CM000162 EDX02139.1 AE014298 BT014631 AAF48957.1 AAT27255.1 AHN59909.1 CH954180 EDV46699.1 CH964291 EDW86428.1 GGMS01014138 MBY83341.1 ABLF02040094 ABLF02040101 ABLF02040106 ABLF02040107 GEBQ01001859 JAT38118.1 CH933812 EDW06119.1 KRG07152.1 CH379063 EAL32546.1 KRT06111.1 CH479193 EDW26910.1 GFXV01006312 MBW18117.1 JXUM01132778 KQ567891 KXJ69234.1 OUUW01000044 SPP89999.1 SPP89997.1 GBBI01002847 JAC15865.1 KK118034 KFM72043.1 KK854884 PTY19479.1 CP012528 ALC49337.1 UFQS01000608 UFQT01000608 SSX05324.1 SSX25685.1 GFDL01012909 JAV22136.1 AXCN02001630 GECZ01023280 JAS46489.1 CH477373 EAT42377.1 GBGD01000390 JAC88499.1 GEDC01013290 GEDC01001201 JAS24008.1 JAS36097.1 GFTR01008278 JAW08148.1 CH940655 EDW66106.1 KRF82563.1 CVRI01000073 CRL07613.1 ABJB010188807 ABJB010509865 ABJB011040524 ABJB011050110 DS642090 EEC01905.1 GDKW01001471 JAI55124.1 CH916376 EDV95303.1 GEZM01010886 JAV93751.1 GDIQ01042247 JAN52490.1 GDIP01135527 JAL68187.1 GDIP01225368 JAI98033.1 GDIP01060938 JAM42777.1 GDIQ01116982 JAL34744.1 GDIQ01028250 JAN66487.1 GANP01000347 JAB84121.1
RVE53059.1 KQ971338 EFA02495.1 GL888187 EGI65507.1 GL764458 EFZ17627.1 KQ414648 KOC66228.1 NNAY01000856 OXU26185.1 GL441439 EFN64828.1 APGK01008041 KB737564 KB632337 ENN83000.1 ERL92856.1 GL449694 EFN82185.1 NEVH01022644 PNF18368.1 LBMM01000733 KMQ97611.1 APGK01009386 KB737932 ENN82834.1 KQ435737 KOX76951.1 AJVK01005172 AJVK01005173 AJVK01005174 ADTU01009482 ADTU01009483 ADTU01009484 KK108118 EZA46879.1 QOIP01000005 RLU22258.1 KQ976476 KYM83848.1 KQ977791 KYM99915.1 KQ982093 KYQ60071.1 KQ979592 KYN20585.1 KQ981727 KYN36699.1 GBYB01003290 GBYB01003291 JAG73057.1 JAG73058.1 GEZM01010884 JAV93754.1 KZ288202 PBC33529.1 GFDF01003184 JAV10900.1 GFDF01003185 JAV10899.1 AJWK01018284 AJWK01018285 AJWK01018286 AJWK01018287 GALX01002292 JAB66174.1 GAMC01006239 GAMC01006238 JAC00318.1 JRES01000940 KNC27009.1 GBXI01007901 JAD06391.1 UFQS01000513 UFQT01000513 SSX04518.1 SSX24881.1 KA645227 AFP59856.1 JXJN01015768 GANO01002031 JAB57840.1 CCAG010002137 GAKP01016707 GAKP01016705 JAC42247.1 DS235078 EEB11459.1 JH431165 GDHF01033695 GDHF01031825 GDHF01027382 GDHF01015353 GDHF01004443 GDHF01002482 GDHF01002480 GDHF01000171 JAI18619.1 JAI20489.1 JAI24932.1 JAI36961.1 JAI47871.1 JAI49832.1 JAI49834.1 JAI52143.1 CH902621 EDV44699.1 GGMR01005361 MBY17980.1 CM000162 EDX02139.1 AE014298 BT014631 AAF48957.1 AAT27255.1 AHN59909.1 CH954180 EDV46699.1 CH964291 EDW86428.1 GGMS01014138 MBY83341.1 ABLF02040094 ABLF02040101 ABLF02040106 ABLF02040107 GEBQ01001859 JAT38118.1 CH933812 EDW06119.1 KRG07152.1 CH379063 EAL32546.1 KRT06111.1 CH479193 EDW26910.1 GFXV01006312 MBW18117.1 JXUM01132778 KQ567891 KXJ69234.1 OUUW01000044 SPP89999.1 SPP89997.1 GBBI01002847 JAC15865.1 KK118034 KFM72043.1 KK854884 PTY19479.1 CP012528 ALC49337.1 UFQS01000608 UFQT01000608 SSX05324.1 SSX25685.1 GFDL01012909 JAV22136.1 AXCN02001630 GECZ01023280 JAS46489.1 CH477373 EAT42377.1 GBGD01000390 JAC88499.1 GEDC01013290 GEDC01001201 JAS24008.1 JAS36097.1 GFTR01008278 JAW08148.1 CH940655 EDW66106.1 KRF82563.1 CVRI01000073 CRL07613.1 ABJB010188807 ABJB010509865 ABJB011040524 ABJB011050110 DS642090 EEC01905.1 GDKW01001471 JAI55124.1 CH916376 EDV95303.1 GEZM01010886 JAV93751.1 GDIQ01042247 JAN52490.1 GDIP01135527 JAL68187.1 GDIP01225368 JAI98033.1 GDIP01060938 JAM42777.1 GDIQ01116982 JAL34744.1 GDIQ01028250 JAN66487.1 GANP01000347 JAB84121.1
Proteomes
UP000005204
UP000283053
UP000007266
UP000007755
UP000053825
UP000215335
+ More
UP000000311 UP000019118 UP000030742 UP000008237 UP000235965 UP000036403 UP000053105 UP000005203 UP000092462 UP000005205 UP000053097 UP000279307 UP000078540 UP000078542 UP000075809 UP000078492 UP000078541 UP000192223 UP000242457 UP000092461 UP000037069 UP000091820 UP000095301 UP000092443 UP000092460 UP000092444 UP000092445 UP000009046 UP000095300 UP000078200 UP000007801 UP000002282 UP000000803 UP000008711 UP000192221 UP000007798 UP000007819 UP000009192 UP000001819 UP000008744 UP000069940 UP000249989 UP000268350 UP000054359 UP000092553 UP000075886 UP000085678 UP000008820 UP000008792 UP000183832 UP000001555 UP000001070 UP000075884
UP000000311 UP000019118 UP000030742 UP000008237 UP000235965 UP000036403 UP000053105 UP000005203 UP000092462 UP000005205 UP000053097 UP000279307 UP000078540 UP000078542 UP000075809 UP000078492 UP000078541 UP000192223 UP000242457 UP000092461 UP000037069 UP000091820 UP000095301 UP000092443 UP000092460 UP000092444 UP000092445 UP000009046 UP000095300 UP000078200 UP000007801 UP000002282 UP000000803 UP000008711 UP000192221 UP000007798 UP000007819 UP000009192 UP000001819 UP000008744 UP000069940 UP000249989 UP000268350 UP000054359 UP000092553 UP000075886 UP000085678 UP000008820 UP000008792 UP000183832 UP000001555 UP000001070 UP000075884
PRIDE
Pfam
Interpro
IPR021183
NatA_aux_su
+ More
IPR013026 TPR-contain_dom
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR024766 Znf_RING_H2
IPR013083 Znf_RING/FYVE/PHD
IPR036291 NAD(P)-bd_dom_sf
IPR008927 6-PGluconate_DH-like_C_sf
IPR006168 G3P_DH_NAD-dep
IPR013328 6PGD_dom2
IPR011128 G3P_DH_NAD-dep_N
IPR006109 G3P_DH_NAD-dep_C
IPR017751 G3P_DH_NAD-dep_euk
IPR013026 TPR-contain_dom
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR024766 Znf_RING_H2
IPR013083 Znf_RING/FYVE/PHD
IPR036291 NAD(P)-bd_dom_sf
IPR008927 6-PGluconate_DH-like_C_sf
IPR006168 G3P_DH_NAD-dep
IPR013328 6PGD_dom2
IPR011128 G3P_DH_NAD-dep_N
IPR006109 G3P_DH_NAD-dep_C
IPR017751 G3P_DH_NAD-dep_euk
Gene 3D
ProteinModelPortal
H9JI91
A0A2W1BZ33
A0A2H1W3K0
A0A437BS50
D2A0B9
F4WJY5
+ More
E9IP70 A0A0L7R5V7 A0A232F6C0 E2ANZ6 N6UMP0 E2BPZ7 A0A2J7PPV0 A0A0J7L4R9 N6TQQ6 A0A0N0U6I2 A0A087ZRP7 A0A1B0FY46 A0A158N9D6 A0A026VVM1 A0A3L8DR34 A0A195BGR2 A0A195CGQ8 A0A151XIC8 A0A195E5W2 A0A195F9H2 A0A0C9R5T2 A0A1Y1N751 A0A1W4XAU5 A0A2A3EQW5 A0A1L8DWR9 A0A1L8DWX1 A0A1B0CM67 V5GPW5 W8BNC4 A0A0L0C6F5 A0A0A1X689 A0A1A9WXX6 A0A336KKR7 T1P942 A0A1I8MMP8 A0A1A9XRZ5 A0A1B0BK08 U5EYF6 A0A1B0G1J1 A0A034VJH4 A0A1A9Z1V2 E0VDK3 A0A1I8NSN7 A0A1A9UXZ8 T1INF4 A0A0K8WFG9 B3MPT7 A0A2S2NLB3 B4PZI3 Q9VWI2 B3NVN4 A0A1W4VTE2 B4NPI8 A0A2S2R005 X1WIP8 A0A1B6MQ90 B4L6X3 Q29IV7 B4GW65 A0A2H8TV72 A0A182HEW7 A0A3B0KTD8 A0A3B0K712 A0A023F378 A0A087U3V4 A0A2R7WIZ7 A0A0M4F9R1 A0A336M640 A0A1Q3F3K0 A0A182QY12 A0A1S3JLG3 A0A1B6F8C2 Q177P1 A0A069DZM3 A0A1B6EDW5 A0A224XED0 A0A1W4XBZ6 B4MA47 A0A1J1J5C4 B7P5N3 A0A0P4VJF5 B4JX27 A0A1Y1N7A8 A0A0P6FVD5 A0A182NG76 A0A0N8CFZ0 A0A0P4YQ39 A0A0P5YCM2 A0A0P5QYP9 A0A0P6HXS5 V5IK41
E9IP70 A0A0L7R5V7 A0A232F6C0 E2ANZ6 N6UMP0 E2BPZ7 A0A2J7PPV0 A0A0J7L4R9 N6TQQ6 A0A0N0U6I2 A0A087ZRP7 A0A1B0FY46 A0A158N9D6 A0A026VVM1 A0A3L8DR34 A0A195BGR2 A0A195CGQ8 A0A151XIC8 A0A195E5W2 A0A195F9H2 A0A0C9R5T2 A0A1Y1N751 A0A1W4XAU5 A0A2A3EQW5 A0A1L8DWR9 A0A1L8DWX1 A0A1B0CM67 V5GPW5 W8BNC4 A0A0L0C6F5 A0A0A1X689 A0A1A9WXX6 A0A336KKR7 T1P942 A0A1I8MMP8 A0A1A9XRZ5 A0A1B0BK08 U5EYF6 A0A1B0G1J1 A0A034VJH4 A0A1A9Z1V2 E0VDK3 A0A1I8NSN7 A0A1A9UXZ8 T1INF4 A0A0K8WFG9 B3MPT7 A0A2S2NLB3 B4PZI3 Q9VWI2 B3NVN4 A0A1W4VTE2 B4NPI8 A0A2S2R005 X1WIP8 A0A1B6MQ90 B4L6X3 Q29IV7 B4GW65 A0A2H8TV72 A0A182HEW7 A0A3B0KTD8 A0A3B0K712 A0A023F378 A0A087U3V4 A0A2R7WIZ7 A0A0M4F9R1 A0A336M640 A0A1Q3F3K0 A0A182QY12 A0A1S3JLG3 A0A1B6F8C2 Q177P1 A0A069DZM3 A0A1B6EDW5 A0A224XED0 A0A1W4XBZ6 B4MA47 A0A1J1J5C4 B7P5N3 A0A0P4VJF5 B4JX27 A0A1Y1N7A8 A0A0P6FVD5 A0A182NG76 A0A0N8CFZ0 A0A0P4YQ39 A0A0P5YCM2 A0A0P5QYP9 A0A0P6HXS5 V5IK41
PDB
6C9M
E-value=1.04319e-36,
Score=383
Ontologies
GO
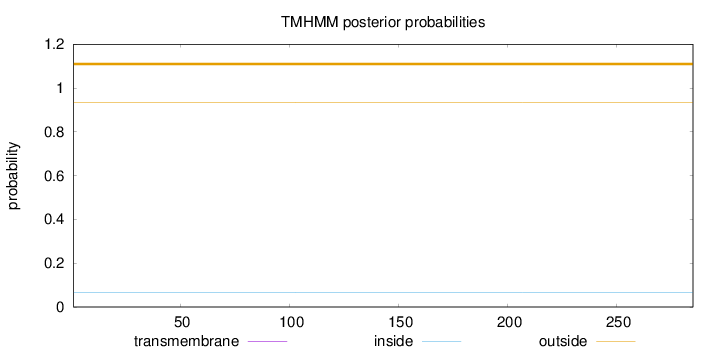
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00052
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06728
outside
1 - 285
Population Genetic Test Statistics
Pi
22.035169
Theta
184.752585
Tajima's D
-1.719647
CLR
1228.963395
CSRT
0.0368981550922454
Interpretation
Possibly Positive selection
