Gene
KWMTBOMO08320
Pre Gene Modal
BGIBMGA009243
Annotation
Uncharacterized_protein_OBRU01_10145_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 2.296
Sequence
CDS
ATGCCACGGTGTATCCAGATCTATTTTCCCAAACATGTCATAAAGCACTCGCTTTCCATCAAATTCTTCTTTCTAGTAAGCTGTTCGAGCCTCTTCCTATCACTAGTTTGGATACAGCTCTACTGTGATACCGAATCGTCCAGGGTCACTGCAAGTCTCGTAGAAGAAGCCCTCGAAAATGTTTCCAAGGATCTCCGTTATTCGGATGTTTATAGGAAGACTAGCAAGCTACCGAAGTTCATCAAGTATATCCTGCTATGGACAAGGCAAGATTTTGCTCCTTTTTATTTCCTCGGGCAAGGTCAACGGGCTTTCCTCAAAAACAATTGCTCGGTGATAAATTGCTACATAACAACTGACAGGAAATTCTTCGGTGGCGATCTCACGAAGTTTGACGGGATAGCGTTCAATGGTCGAAATATGAAAAGTTCAGATTTGCCGAGAATCCGTACGGGAAAACAGAAATATGTGTTCTTCAACATGGAGTCTGCGGAAAATTTTCCCGTTTGCAGCGAAAAGTTCGATGGGTTTTTCAATTGGACAGCCACTTATAGACTGGATTCAGATATTCCGTTCCCATATATACAAATAAGAAATATTAATGGTGAGATCGTTGGTCCGAAGAAAGAAATGGTATGGGCGGACGACTTTTCGGAGATTGATGAAGATCTGTCCTTGAAAATTGCCAATAAGACTAAAGCAGCTGCTTGGTTCGTCTCTAATTGTAAGAGTAAAAGCGGACGAAAGAAATTTGTGGATGATTTACAAAGGGCCTTAAATCGATTTAAATATTCTATCGATATTTACGGATCGTGTGGAAAGTTAAAATGTCCAAGGGACAAGAATAACGATTGTCATTCGCTGTTGGAAAAAGATTACTATTTTTATTTATCGTTAGAGAATACGTTTGCGGAAGATTATGTCACAGAGAAGTTGTTAACTGCATTACAGCATGACGTAGTGCCGATAGTGTATGGCGGAGCGAATTATTCTAGGTTTCTTCCACCTGGCTCTTACATCGACGGGAGAAGATACAAGCCCGCTCAACTTGCGGAAATTATGAACAAACTCATGAAAAATCACAAGTTGTACGCCGAATATTTCCGTTGGAAGAAGCACTACACGTACCACGACCCTTCGGAGGTGGACAACGTTTGTGCTCTCTGTAAGGCATTGAATAACAAAAAAATGGCCAGTCCCAATACTTATTATCAGTTCAGGAAGTGGTGGCATCCAGAATTCAAGAGATTGTGTTAG
Protein
MPRCIQIYFPKHVIKHSLSIKFFFLVSCSSLFLSLVWIQLYCDTESSRVTASLVEEALENVSKDLRYSDVYRKTSKLPKFIKYILLWTRQDFAPFYFLGQGQRAFLKNNCSVINCYITTDRKFFGGDLTKFDGIAFNGRNMKSSDLPRIRTGKQKYVFFNMESAENFPVCSEKFDGFFNWTATYRLDSDIPFPYIQIRNINGEIVGPKKEMVWADDFSEIDEDLSLKIANKTKAAAWFVSNCKSKSGRKKFVDDLQRALNRFKYSIDIYGSCGKLKCPRDKNNDCHSLLEKDYYFYLSLENTFAEDYVTEKLLTALQHDVVPIVYGGANYSRFLPPGSYIDGRRYKPAQLAEIMNKLMKNHKLYAEYFRWKKHYTYHDPSEVDNVCALCKALNNKKMASPNTYYQFRKWWHPEFKRLC
Summary
Similarity
Belongs to the glycosyltransferase 10 family.
Uniprot
H9JI93
A0A0L7LBT7
A0A2W1BS87
A0A2H1V9M8
A0A0L7LD09
A0A212EZY5
+ More
A0A3S2LG40 A0A0L7KT82 A0A2A4J121 A0A194R6N8 A0A194R740 A0A194Q472 A0A194QTG1 A0A212F4A2 A0A2H1VKP0 A0A2A4J733 A0A212FDW4 A0A437BN03 A0A3S2NPV0 A0A3S2LHN9 A0A0L7KKB0 A0A0L7K4N4 A0A3S2M564 A0A2H1X3C4 A0A2A4J5I4 A0A437BMJ2 A0A2H1VN94 A0A3S2PHJ4 A0A3S2NI47 A0A194QV22 A0A3S2NK67 A0A194PTI0 A0A2W1BWB4 A0A2A4J738 A0A2W1C1F3 A0A2H1WHJ5 H9JI96 A0A3S2PJA5 A0A212F5Z8 A0A2W1BGR4 A0A2A4J2N7 A0A2A4JYW9 A0A212FHM7 A0A3S2NZG0 A0A2H1VRA0 A0A2W1BZ24 A0A0L7KR78 A0A3S2M779 A0A2H1VCS8 A0A310SMV6 A0A2A4J0X4 A0A2H1V6K5 A0A0L7RBK3 A0A2A3ET90 Q05GU1 A0A088AJ84 A0A0L7KUY7 A0A154PNF0 A0A0P5UE00 A0A0P6CB97 A0A0P5S2V2 A0A0P5UR34 A0A164YS43 A0A0P6HZP4 A0A0P6EHA9 A0A0P6IKS2 A0A0P5UXD4 A0A084WRX7 A0A182NZS3 A0A0P5KLH8 A0A0P5NPN0 A0A0P6ASP9 A0A182XK10 A0A084WRX8 A0A0P6J2W8 A0A158P137 E9IYL6 A0A182IAZ0 Q7QF91 A0A0P5I7R2 A0A182V5C9 A0A0N8DRT3 A0A182FLK0 A0A182KFG3 E2BU28 A0A182UEH9 A0A151K0V3 A0A0N8C4Z8 A0A0P6E219 A0A0P5DYJ8
A0A3S2LG40 A0A0L7KT82 A0A2A4J121 A0A194R6N8 A0A194R740 A0A194Q472 A0A194QTG1 A0A212F4A2 A0A2H1VKP0 A0A2A4J733 A0A212FDW4 A0A437BN03 A0A3S2NPV0 A0A3S2LHN9 A0A0L7KKB0 A0A0L7K4N4 A0A3S2M564 A0A2H1X3C4 A0A2A4J5I4 A0A437BMJ2 A0A2H1VN94 A0A3S2PHJ4 A0A3S2NI47 A0A194QV22 A0A3S2NK67 A0A194PTI0 A0A2W1BWB4 A0A2A4J738 A0A2W1C1F3 A0A2H1WHJ5 H9JI96 A0A3S2PJA5 A0A212F5Z8 A0A2W1BGR4 A0A2A4J2N7 A0A2A4JYW9 A0A212FHM7 A0A3S2NZG0 A0A2H1VRA0 A0A2W1BZ24 A0A0L7KR78 A0A3S2M779 A0A2H1VCS8 A0A310SMV6 A0A2A4J0X4 A0A2H1V6K5 A0A0L7RBK3 A0A2A3ET90 Q05GU1 A0A088AJ84 A0A0L7KUY7 A0A154PNF0 A0A0P5UE00 A0A0P6CB97 A0A0P5S2V2 A0A0P5UR34 A0A164YS43 A0A0P6HZP4 A0A0P6EHA9 A0A0P6IKS2 A0A0P5UXD4 A0A084WRX7 A0A182NZS3 A0A0P5KLH8 A0A0P5NPN0 A0A0P6ASP9 A0A182XK10 A0A084WRX8 A0A0P6J2W8 A0A158P137 E9IYL6 A0A182IAZ0 Q7QF91 A0A0P5I7R2 A0A182V5C9 A0A0N8DRT3 A0A182FLK0 A0A182KFG3 E2BU28 A0A182UEH9 A0A151K0V3 A0A0N8C4Z8 A0A0P6E219 A0A0P5DYJ8
Pubmed
EMBL
BABH01022242
JTDY01001793
KOB72859.1
KZ149916
PZC77942.1
ODYU01001405
+ More
SOQ37538.1 JTDY01001718 KOB73086.1 AGBW02011221 OWR47017.1 RSAL01000016 RVE53056.1 JTDY01005919 KOB66482.1 NWSH01004327 PCG65224.1 KQ460627 KPJ13478.1 KPJ13477.1 KQ459585 KPI98210.1 KQ461137 KPJ08813.1 AGBW02010417 OWR48543.1 ODYU01003079 SOQ41387.1 NWSH01002952 PCG67232.1 AGBW02008995 OWR51946.1 RSAL01000031 RVE51638.1 RSAL01000164 RVE45329.1 RSAL01000116 RVE46859.1 JTDY01009552 KOB63349.1 JTDY01010816 KOB57987.1 RVE51643.1 ODYU01013118 SOQ59805.1 PCG67231.1 RVE51644.1 ODYU01003471 SOQ42305.1 RVE51641.1 RVE51642.1 KQ461108 KPJ09317.1 RVE53051.1 KQ459594 KPI96064.1 PZC77935.1 PCG67233.1 PZC77933.1 ODYU01008712 SOQ52539.1 BABH01022215 RVE53050.1 AGBW02010111 OWR49153.1 KZ150145 PZC72905.1 NWSH01003936 PCG65653.1 NWSH01000372 PCG76878.1 AGBW02008491 OWR53234.1 RVE53053.1 ODYU01003968 SOQ43373.1 PZC77936.1 JTDY01006679 KOB65783.1 RVE53052.1 ODYU01001860 SOQ38649.1 KQ761844 OAD56586.1 NWSH01004476 PCG65062.1 ODYU01000952 SOQ36485.1 KQ414617 KOC68244.1 KZ288185 PBC34965.1 AM279413 CAK50261.1 JTDY01005492 KOB66945.1 KQ434977 KZC12838.1 GDIP01114408 JAL89306.1 GDIP01002675 JAN01041.1 GDIQ01093775 JAL57951.1 GDIP01109554 JAL94160.1 LRGB01000868 KZS15541.1 GDIQ01012846 JAN81891.1 GDIQ01076840 JAN17897.1 GDIQ01002877 JAN91860.1 GDIP01107297 JAL96417.1 ATLV01026273 KE525409 KFB52971.1 GDIQ01183544 JAK68181.1 GDIQ01146178 JAL05548.1 GDIP01038939 JAM64776.1 KFB52972.1 GDIQ01067914 GDIQ01013905 JAN80832.1 ADTU01005498 ADTU01005499 ADTU01005500 GL767018 EFZ14350.1 APCN01001363 AAAB01008846 EAA06326.5 GDIQ01217631 JAK34094.1 GDIP01007183 JAM96532.1 GL450575 EFN80802.1 KQ981250 KYN44218.1 GDIQ01114275 JAL37451.1 GDIQ01083834 JAN10903.1 GDIP01167237 JAJ56165.1
SOQ37538.1 JTDY01001718 KOB73086.1 AGBW02011221 OWR47017.1 RSAL01000016 RVE53056.1 JTDY01005919 KOB66482.1 NWSH01004327 PCG65224.1 KQ460627 KPJ13478.1 KPJ13477.1 KQ459585 KPI98210.1 KQ461137 KPJ08813.1 AGBW02010417 OWR48543.1 ODYU01003079 SOQ41387.1 NWSH01002952 PCG67232.1 AGBW02008995 OWR51946.1 RSAL01000031 RVE51638.1 RSAL01000164 RVE45329.1 RSAL01000116 RVE46859.1 JTDY01009552 KOB63349.1 JTDY01010816 KOB57987.1 RVE51643.1 ODYU01013118 SOQ59805.1 PCG67231.1 RVE51644.1 ODYU01003471 SOQ42305.1 RVE51641.1 RVE51642.1 KQ461108 KPJ09317.1 RVE53051.1 KQ459594 KPI96064.1 PZC77935.1 PCG67233.1 PZC77933.1 ODYU01008712 SOQ52539.1 BABH01022215 RVE53050.1 AGBW02010111 OWR49153.1 KZ150145 PZC72905.1 NWSH01003936 PCG65653.1 NWSH01000372 PCG76878.1 AGBW02008491 OWR53234.1 RVE53053.1 ODYU01003968 SOQ43373.1 PZC77936.1 JTDY01006679 KOB65783.1 RVE53052.1 ODYU01001860 SOQ38649.1 KQ761844 OAD56586.1 NWSH01004476 PCG65062.1 ODYU01000952 SOQ36485.1 KQ414617 KOC68244.1 KZ288185 PBC34965.1 AM279413 CAK50261.1 JTDY01005492 KOB66945.1 KQ434977 KZC12838.1 GDIP01114408 JAL89306.1 GDIP01002675 JAN01041.1 GDIQ01093775 JAL57951.1 GDIP01109554 JAL94160.1 LRGB01000868 KZS15541.1 GDIQ01012846 JAN81891.1 GDIQ01076840 JAN17897.1 GDIQ01002877 JAN91860.1 GDIP01107297 JAL96417.1 ATLV01026273 KE525409 KFB52971.1 GDIQ01183544 JAK68181.1 GDIQ01146178 JAL05548.1 GDIP01038939 JAM64776.1 KFB52972.1 GDIQ01067914 GDIQ01013905 JAN80832.1 ADTU01005498 ADTU01005499 ADTU01005500 GL767018 EFZ14350.1 APCN01001363 AAAB01008846 EAA06326.5 GDIQ01217631 JAK34094.1 GDIP01007183 JAM96532.1 GL450575 EFN80802.1 KQ981250 KYN44218.1 GDIQ01114275 JAL37451.1 GDIQ01083834 JAN10903.1 GDIP01167237 JAJ56165.1
Proteomes
Gene 3D
ProteinModelPortal
H9JI93
A0A0L7LBT7
A0A2W1BS87
A0A2H1V9M8
A0A0L7LD09
A0A212EZY5
+ More
A0A3S2LG40 A0A0L7KT82 A0A2A4J121 A0A194R6N8 A0A194R740 A0A194Q472 A0A194QTG1 A0A212F4A2 A0A2H1VKP0 A0A2A4J733 A0A212FDW4 A0A437BN03 A0A3S2NPV0 A0A3S2LHN9 A0A0L7KKB0 A0A0L7K4N4 A0A3S2M564 A0A2H1X3C4 A0A2A4J5I4 A0A437BMJ2 A0A2H1VN94 A0A3S2PHJ4 A0A3S2NI47 A0A194QV22 A0A3S2NK67 A0A194PTI0 A0A2W1BWB4 A0A2A4J738 A0A2W1C1F3 A0A2H1WHJ5 H9JI96 A0A3S2PJA5 A0A212F5Z8 A0A2W1BGR4 A0A2A4J2N7 A0A2A4JYW9 A0A212FHM7 A0A3S2NZG0 A0A2H1VRA0 A0A2W1BZ24 A0A0L7KR78 A0A3S2M779 A0A2H1VCS8 A0A310SMV6 A0A2A4J0X4 A0A2H1V6K5 A0A0L7RBK3 A0A2A3ET90 Q05GU1 A0A088AJ84 A0A0L7KUY7 A0A154PNF0 A0A0P5UE00 A0A0P6CB97 A0A0P5S2V2 A0A0P5UR34 A0A164YS43 A0A0P6HZP4 A0A0P6EHA9 A0A0P6IKS2 A0A0P5UXD4 A0A084WRX7 A0A182NZS3 A0A0P5KLH8 A0A0P5NPN0 A0A0P6ASP9 A0A182XK10 A0A084WRX8 A0A0P6J2W8 A0A158P137 E9IYL6 A0A182IAZ0 Q7QF91 A0A0P5I7R2 A0A182V5C9 A0A0N8DRT3 A0A182FLK0 A0A182KFG3 E2BU28 A0A182UEH9 A0A151K0V3 A0A0N8C4Z8 A0A0P6E219 A0A0P5DYJ8
A0A3S2LG40 A0A0L7KT82 A0A2A4J121 A0A194R6N8 A0A194R740 A0A194Q472 A0A194QTG1 A0A212F4A2 A0A2H1VKP0 A0A2A4J733 A0A212FDW4 A0A437BN03 A0A3S2NPV0 A0A3S2LHN9 A0A0L7KKB0 A0A0L7K4N4 A0A3S2M564 A0A2H1X3C4 A0A2A4J5I4 A0A437BMJ2 A0A2H1VN94 A0A3S2PHJ4 A0A3S2NI47 A0A194QV22 A0A3S2NK67 A0A194PTI0 A0A2W1BWB4 A0A2A4J738 A0A2W1C1F3 A0A2H1WHJ5 H9JI96 A0A3S2PJA5 A0A212F5Z8 A0A2W1BGR4 A0A2A4J2N7 A0A2A4JYW9 A0A212FHM7 A0A3S2NZG0 A0A2H1VRA0 A0A2W1BZ24 A0A0L7KR78 A0A3S2M779 A0A2H1VCS8 A0A310SMV6 A0A2A4J0X4 A0A2H1V6K5 A0A0L7RBK3 A0A2A3ET90 Q05GU1 A0A088AJ84 A0A0L7KUY7 A0A154PNF0 A0A0P5UE00 A0A0P6CB97 A0A0P5S2V2 A0A0P5UR34 A0A164YS43 A0A0P6HZP4 A0A0P6EHA9 A0A0P6IKS2 A0A0P5UXD4 A0A084WRX7 A0A182NZS3 A0A0P5KLH8 A0A0P5NPN0 A0A0P6ASP9 A0A182XK10 A0A084WRX8 A0A0P6J2W8 A0A158P137 E9IYL6 A0A182IAZ0 Q7QF91 A0A0P5I7R2 A0A182V5C9 A0A0N8DRT3 A0A182FLK0 A0A182KFG3 E2BU28 A0A182UEH9 A0A151K0V3 A0A0N8C4Z8 A0A0P6E219 A0A0P5DYJ8
PDB
2NZY
E-value=0.0717662,
Score=84
Ontologies
GO
PANTHER
Topology
Subcellular location
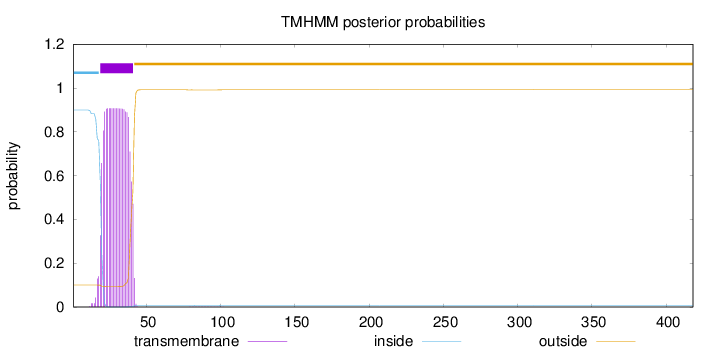
Length:
418
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.40686
Exp number, first 60 AAs:
19.38091
Total prob of N-in:
0.89952
POSSIBLE N-term signal
sequence
inside
1 - 18
TMhelix
19 - 41
outside
42 - 418
Population Genetic Test Statistics
Pi
169.017654
Theta
151.384066
Tajima's D
0.341131
CLR
278.906527
CSRT
0.462626868656567
Interpretation
Uncertain
