Gene
KWMTBOMO08319
Pre Gene Modal
BGIBMGA011165
Annotation
PREDICTED:_uncharacterized_protein_LOC105391048_[Plutella_xylostella]
Full name
Galactose-1-phosphate uridylyltransferase
Location in the cell
Nuclear Reliability : 2.412
Sequence
CDS
ATGAGGCTTGTATGGTACTCGTGTCTTGCCCTTTTCGGTTTGATTACAGCTAAAAAATTTGGTTGTGACAAGAATCTGGATCCTGCCGATGATCCGAAAAACATCGAAAGACCTTGTCCGAACTTCGACTTAGACTGTATTCGAGAATATTTCTCTAGTAATTCAAAATGTCAAATAGCACGGGAGCCAGTACCGGATCCACTGCTTCTCAACCATTACAGATTAAACATAGCAAATTCTAATATAACTATGGAACTCAATAATGTAGAAGTCGGAGGTCTTAATGGAAATATTGTTGAGTTTTATATAAACAAAGAAACTCAGAAACTGGTATTGGTAATAGAAGTGGACTCTGTAAGATTGGAATCGATACAAGTAACATTCAAGTACAATAGAAAGGGTAAAGAGCCCATTGTGAGAAGTGATAATGCCACGTGTGATTACGGACCTATGAGTTTTACTGTGGTGTTTCCGAGCATCTGCGATCTTGATTTGAACAACGCCGAAGTTTTCACGTACATTGAGGATTGTAACACGAAATATACAATGGGAATAGGTCTATATTCCAGTTTGGATCCAAAGATGTCTCAAGAGATCTACAGAGTTGGTGTGCGCATTCCGCCATTCTGCTCAGAGGACCCAGAAATGTGGTTCAGTCAGATAGAGATGCAATTTAAGCTCTCAGGGATATCTGTGGACGATACTAAGTTTACGTATGTGTCAGCGAACCTGGAACCACAGTACGCCAAGGAGGTAAGAGACATCATAACGAAACCGCCAGCTTCAAATAAATACGAAAAACTCAAAACAGAGCTGATCAAGCGACTATCAATATCACAGGAGAAGCGTGTAAGACAACTTCTCATGCACGAAGAACTCGGAGATCGGAAACCGTCTCAGTTTCTACGCCACTTGACAAGTCTAGCTGGTTCAGACGTTCCATCTGATTTTATAATAACTATCTGGACCAGTCGTCTGCCTGTGAACGTGCAAACAGCAGTCGCTACGATTAAAGGATCGTCAATGGAGAAGCTAAGTGACTTGGCAGATGTCGTATATGAAATGGCCCCAACAGTACCACAGGTGGCAAGCGCGTCTTCCCAAGCCCAAGGCAATGAGCAAGGCAATCCGACGATGACTGAGATGGCCCAGCAAATCAATTCGCTGACACAGCAAATCAGCATGCTCTCTAAACAGTTTCGCGTCGGAAAACTCCTGGGGCGGTCGGAAGTAGCGGCCAACGACTGCCCACATTCTTCAGGCCGTCTTTTTATCACCGACCGTACTTCAAAAATGCAGTTCCTAGTGGACACTGGTTCCGATTTGTGTGTTTTTCCACGGTCGGCTGTGAAGGCGCCCTGTACCAAGTCGAAATATGATCTCGTAGCCGCGAATAATTCTATTATACACACCTATGGCCCTGTAACTCTTCATTTAGACTTTGGACTAAGAAGAGACTTCGCGTGGAAATTCACTGTTGCAGACGTATCTAAGCCCATAATCGGAGTGGATTTCTTGTCTTACTACAACTTACTCGTCGATTGCAGAAAACAACGTATTATTGATGGCATCACTTCTCTGTCTGCAGTCGCGACACCTATCGGGATGACTAAAGACCTTCCTTCTGTCAAAGCCGTCACTGGTGAAACTGACTTCCACGATATTCTGAGAGAGTTCCCAGAAATCACAAGGCCAGCAGGTATGCTCTCTACCACATTTAAGCATAACACCGTTCATCATATTAGAACTACACCGGGACCTCCCGTTGCTTGCAAGCCTCGACGTTTGGATCCAGCACGTATGCAAGCCGCGAAGAAAGAGTTCGAGGATATGTTGGCAAGCGGCGTAGCCCAGCGTTCAGAGAGTGCATGGTCTTCTGCTCTGCACTTGGTAAAGAAGAAAGATGACGGATGGCGTCCTTGTGGCGATTACCGATCACTGAATGCGAGAACGATACCTGATAGATATCCAGTACGCCACATCCATGACTTCTCACAACAGGTTTCTGGATGTTCCATCTTTTCTAAAATTGATTTGGTGAAGGCTTATCATCAGATAGCCGTTAATCCTGAGGATATACCTAAAACAGCCATCACTACTCCTTTTGGACTTTTCGAATTTCCATTTATGACCTTTAGTTTGAGGAATGCCGCTCAGACATTCCAAAGGTTCATAGATGAGGTGCTACTAGGCCTCGACTTCACCTATGGATATATTGATGATATATTACTGTTTTCTTCATCGCTAGAGGAACATAAGTGTCATCTACGTCAACTCTTTGATCGTCTCCGGCAGTATGGCATTCTGGTGAACACAACTAAATGTACTTTTGGTCAATCTGTCGTGGAATTCCTAGGATATGAAGTATCTGCTACAGGTACTAAGCCATTGGACACTAAAGTCCAAGCTATCCAGGAATTTCCTGCTCCTAAAACGGTGAAGGAACTCCGAAGATTTTTAGGCATGCTTAATTTCTACCGCCGTTTCATACCTAACGCCGCCCAGTTACAAGCTCCGTTGAACAACGCTCTGGCAGGAGTTAAAATGAAAGGTTCACACCCTGTGGATATGGATGCAGTCATGTTAAAAGCCTTTGAGGACTGTAAGACTTCTTTATCTCAGGCAACGCTTTTGTCTCATCCGGATCCAACCGCCGATCTTGCGATTGTCACTGACGCGTCTCTCACAGCGATTGGAGCTGTTCTACAACAAAGGAAAGGTGAACATTGGGAACCACTCGCTTTCTACTCCCACAAGCTTAGTTCCACCCAATCGAAGTACAGTCCATATGATCGAGAATTACTGTCAGTTTATGAGGCAATAAAGCACTTTCGACACATGATTGAAGCGAGAAATTTCACCGTCTTCACTGATCATAAACCGCTGATCTTTGCCTTCTCAGTAAACAGGGAGAAATGTTCACCAAGGCAATTCAGATATCTTGACTACATATCACAGTTTACTACGGATATTAAATATATACCTGGAAAGGATAACGTAGTCGCCGACGCACTTTCTCGGGTGGATGAAGTCGCTCCATGTATCGACTATGAACATCTTGCACGATCTCAGGAACAAGATCCAGAGCTGCAAGATCTTCTGCGTAATGGAAGTTCTCTAAAATTAGAAAAGTTATCTTCTGAGTATAGTAGCTTACCCGTTTATTGCGACACCAGTGTGCACCCTCCACGACCATTCATCACAAAAGATATGCGAAGACAAGTCTTCAACACACTACATGGACTTCATCACCCAGGATCGTCCGCTACAGTCCGACTGGTGGCGCAAAGATTTGTGTGGCCAGGGATTCGGAAAGACTGTAGAGAGTGGTCAAGACAATGCTTAAACTGTCAGAGAAGTAAAGTAAGTCGCCATACAGTTTCTCCTCTTTCCTCTTTCCTTACCCCTTCGTCAAGATTCACTCATATCCATCTTGACATCGTAGGGCCTCTAAATCTGTCTGCTGGTTTCAGGTACTGCCTCACTGTCATTGACCGCTTCACCCGATGGCCTGAGGTGATACCACTCAGCGAAATTACAGCTGAAGCTTGTGCTGCCGCTCTCGTTAGCGGCTGGATTGCCAGATTTGGATGTCCACTGAGAGTAACTACCGACCGCGGTCGTCAATTCGAGAGTCACTTATTCAAAGCCATAGCTGCATTAATAGGTGCTCACCACCTCCGGACTACCGCATATCACCCCGCCGCCAACGGAATAGTCGAACGAATGCATCGCCAGTTGAAGGCAGCCATCATGTGCCATACACCTTCGCAATGGACTGAAGCACTTCCTCTCGTACTGATGGGCATGCGCAGTTCCTGGAAGGATGACATTGACGCATCTCCTGCAGAGCTGGTGTACGGCGAGCCCCTCTGTCTGCCAGGTCAATTCATTTCACCAAATGAAGACTACAAAGTGTCTGACGTCACCCAGTATGCAACTCGTCTACGTGCATGCATGGCGAATTTGACACCGAGACCGACATCCTGGCATAGCAATTCACCATTTTATATACCTCGCGATTTACATACAACCTCCCATGTCTTCATCCGACAAGATCATGTGCGTGGTTCTCTGGTACCACCATATGCTGGTCCCTTCAAGGTTGTCAAAAGGCAGCCAAAATTCTTCACTGTCAAAGTTCGAGGGAAGCTTGTAAATGTCTCCATAGATCGTCTAAAGCCGGCCTATATGACGAGAGTTCCAGAAGATCGTGTAGCCGAAAGAGAGTATCAGAAACTGATAGCTTCTATGCCAGGTGCCGTTCTAGAAAGTTTTATCACGGAAGGAAAAAAATTCATGGACCATTACATACAATACAATATCTGCAACTTTGGATTAGAATTAGGCTGTGAATAA
Protein
MRLVWYSCLALFGLITAKKFGCDKNLDPADDPKNIERPCPNFDLDCIREYFSSNSKCQIAREPVPDPLLLNHYRLNIANSNITMELNNVEVGGLNGNIVEFYINKETQKLVLVIEVDSVRLESIQVTFKYNRKGKEPIVRSDNATCDYGPMSFTVVFPSICDLDLNNAEVFTYIEDCNTKYTMGIGLYSSLDPKMSQEIYRVGVRIPPFCSEDPEMWFSQIEMQFKLSGISVDDTKFTYVSANLEPQYAKEVRDIITKPPASNKYEKLKTELIKRLSISQEKRVRQLLMHEELGDRKPSQFLRHLTSLAGSDVPSDFIITIWTSRLPVNVQTAVATIKGSSMEKLSDLADVVYEMAPTVPQVASASSQAQGNEQGNPTMTEMAQQINSLTQQISMLSKQFRVGKLLGRSEVAANDCPHSSGRLFITDRTSKMQFLVDTGSDLCVFPRSAVKAPCTKSKYDLVAANNSIIHTYGPVTLHLDFGLRRDFAWKFTVADVSKPIIGVDFLSYYNLLVDCRKQRIIDGITSLSAVATPIGMTKDLPSVKAVTGETDFHDILREFPEITRPAGMLSTTFKHNTVHHIRTTPGPPVACKPRRLDPARMQAAKKEFEDMLASGVAQRSESAWSSALHLVKKKDDGWRPCGDYRSLNARTIPDRYPVRHIHDFSQQVSGCSIFSKIDLVKAYHQIAVNPEDIPKTAITTPFGLFEFPFMTFSLRNAAQTFQRFIDEVLLGLDFTYGYIDDILLFSSSLEEHKCHLRQLFDRLRQYGILVNTTKCTFGQSVVEFLGYEVSATGTKPLDTKVQAIQEFPAPKTVKELRRFLGMLNFYRRFIPNAAQLQAPLNNALAGVKMKGSHPVDMDAVMLKAFEDCKTSLSQATLLSHPDPTADLAIVTDASLTAIGAVLQQRKGEHWEPLAFYSHKLSSTQSKYSPYDRELLSVYEAIKHFRHMIEARNFTVFTDHKPLIFAFSVNREKCSPRQFRYLDYISQFTTDIKYIPGKDNVVADALSRVDEVAPCIDYEHLARSQEQDPELQDLLRNGSSLKLEKLSSEYSSLPVYCDTSVHPPRPFITKDMRRQVFNTLHGLHHPGSSATVRLVAQRFVWPGIRKDCREWSRQCLNCQRSKVSRHTVSPLSSFLTPSSRFTHIHLDIVGPLNLSAGFRYCLTVIDRFTRWPEVIPLSEITAEACAAALVSGWIARFGCPLRVTTDRGRQFESHLFKAIAALIGAHHLRTTAYHPAANGIVERMHRQLKAAIMCHTPSQWTEALPLVLMGMRSSWKDDIDASPAELVYGEPLCLPGQFISPNEDYKVSDVTQYATRLRACMANLTPRPTSWHSNSPFYIPRDLHTTSHVFIRQDHVRGSLVPPYAGPFKVVKRQPKFFTVKVRGKLVNVSIDRLKPAYMTRVPEDRVAEREYQKLIASMPGAVLESFITEGKKFMDHYIQYNICNFGLELGCE
Summary
Catalytic Activity
alpha-D-galactose 1-phosphate + UDP-alpha-D-glucose = alpha-D-glucose 1-phosphate + UDP-alpha-D-galactose
Similarity
Belongs to the galactose-1-phosphate uridylyltransferase type 1 family.
Belongs to the histone H3 family.
Belongs to the histone H3 family.
Uniprot
A0A0J7NWH7
D6WTN9
W4Y2D0
A0A085M208
W4XB88
J9KB96
+ More
W4Y5A8 A0A1X7VKM7 W4YUC1 Q9BM81 G7Y855 Q9BM79 Q9BM80 A0A0J7K7I7 W4ZBK7 C7C1Z9 A0A085MR14 A0A147BLP1 A0A085N6R2 A0A085N6P6 A0A085N5A1 A0A0C2MTE2 A0A0C2N259 X1XPJ2 X1XU40 A0A085MTV7 A0A0C2NBN0 A0A0C2MJW7 A0A0C2NDW5 X1XTW4 A0A085NRB3 K7J8H5 A0A085LN63 X1X9N5 A0A0V1F272 A0A085N387 L7LZ63 K7JCJ0 A0A147BJG9 X1XU43 K7JAG6 A0A0V0TCH2 A0A0V1FLR3 A0A085LKW1 A0A085MDV5 X1XU27 K7ISS4 A0A085LRR2 C7C1Z7 K7JBH7 A0A0V1KEE1 J9K9L2
W4Y5A8 A0A1X7VKM7 W4YUC1 Q9BM81 G7Y855 Q9BM79 Q9BM80 A0A0J7K7I7 W4ZBK7 C7C1Z9 A0A085MR14 A0A147BLP1 A0A085N6R2 A0A085N6P6 A0A085N5A1 A0A0C2MTE2 A0A0C2N259 X1XPJ2 X1XU40 A0A085MTV7 A0A0C2NBN0 A0A0C2MJW7 A0A0C2NDW5 X1XTW4 A0A085NRB3 K7J8H5 A0A085LN63 X1X9N5 A0A0V1F272 A0A085N387 L7LZ63 K7JCJ0 A0A147BJG9 X1XU43 K7JAG6 A0A0V0TCH2 A0A0V1FLR3 A0A085LKW1 A0A085MDV5 X1XU27 K7ISS4 A0A085LRR2 C7C1Z7 K7JBH7 A0A0V1KEE1 J9K9L2
EC Number
2.7.7.12
EMBL
LBMM01001181
KMQ96765.1
KQ971355
EFA07205.2
AAGJ04130731
KL363241
+ More
KFD51254.1 AAGJ04001742 ABLF02030693 ABLF02030697 AAGJ04112098 AAGJ04172791 AY013563 AAK07485.1 DF142933 GAA49140.1 AY013569 AAK07487.1 AY013565 AAK07486.1 LBMM01012567 KMQ86146.1 AAGJ04000250 FN356213 CAX83702.1 KL367778 KFD59660.1 GEGO01003687 JAR91717.1 KL367543 KFD65158.1 KL367544 KFD65142.1 KL367552 KFD64647.1 JWZT01003175 KII67485.1 JWZT01002058 KII70465.1 ABLF02058233 ABLF02040931 ABLF02040938 KL367655 KFD60653.1 JWZT01005177 JWZT01001778 KII61927.1 KII71382.1 JWZT01004124 KII64665.1 JWZT01001363 KII72172.1 ABLF02005645 ABLF02042063 KL367479 KFD72009.1 AAZX01013940 KL363373 KFD46409.1 JYDR01000002 KRY79251.1 KL367564 KFD63933.1 GACK01008900 JAA56134.1 AAZX01006801 AAZX01023483 GEGO01004475 JAR90929.1 ABLF02006352 ABLF02013150 ABLF02013854 ABLF02041505 AAZX01023279 JYDJ01000353 KRX36608.1 JYDT01000067 KRY86703.1 KL363454 KFD45607.1 KL363200 KFD55401.1 ABLF02057446 ABLF02059301 AAZX01009606 KL363318 KFD47658.1 FN356212 CAX83701.1 AAZX01023264 JYDV01000002 KRZ45603.1 ABLF02011086 ABLF02042879
KFD51254.1 AAGJ04001742 ABLF02030693 ABLF02030697 AAGJ04112098 AAGJ04172791 AY013563 AAK07485.1 DF142933 GAA49140.1 AY013569 AAK07487.1 AY013565 AAK07486.1 LBMM01012567 KMQ86146.1 AAGJ04000250 FN356213 CAX83702.1 KL367778 KFD59660.1 GEGO01003687 JAR91717.1 KL367543 KFD65158.1 KL367544 KFD65142.1 KL367552 KFD64647.1 JWZT01003175 KII67485.1 JWZT01002058 KII70465.1 ABLF02058233 ABLF02040931 ABLF02040938 KL367655 KFD60653.1 JWZT01005177 JWZT01001778 KII61927.1 KII71382.1 JWZT01004124 KII64665.1 JWZT01001363 KII72172.1 ABLF02005645 ABLF02042063 KL367479 KFD72009.1 AAZX01013940 KL363373 KFD46409.1 JYDR01000002 KRY79251.1 KL367564 KFD63933.1 GACK01008900 JAA56134.1 AAZX01006801 AAZX01023483 GEGO01004475 JAR90929.1 ABLF02006352 ABLF02013150 ABLF02013854 ABLF02041505 AAZX01023279 JYDJ01000353 KRX36608.1 JYDT01000067 KRY86703.1 KL363454 KFD45607.1 KL363200 KFD55401.1 ABLF02057446 ABLF02059301 AAZX01009606 KL363318 KFD47658.1 FN356212 CAX83701.1 AAZX01023264 JYDV01000002 KRZ45603.1 ABLF02011086 ABLF02042879
Proteomes
PRIDE
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR000477 RT_dom
IPR034132 RP_Saci-like
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR005849 GalP_Utransf_N
IPR001969 Aspartic_peptidase_AS
IPR019779 GalP_UDPtransf1_His-AS
IPR001995 Peptidase_A2_cat
IPR001937 GalP_UDPtransf1
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR036265 HIT-like_sf
IPR041373 RT_RNaseH
IPR016024 ARM-type_fold
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR008383 API5
IPR038718 SNF2-like_sf
IPR027417 P-loop_NTPase
IPR000330 SNF2_N
IPR027806 HARBI1_dom
IPR000164 Histone_H3/CENP-A
IPR002119 Histone_H2A
IPR032454 Histone_H2A_C
IPR007125 Histone_H2A/H2B/H3
IPR009072 Histone-fold
IPR018061 Retropepsins
IPR000477 RT_dom
IPR034132 RP_Saci-like
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR005849 GalP_Utransf_N
IPR001969 Aspartic_peptidase_AS
IPR019779 GalP_UDPtransf1_His-AS
IPR001995 Peptidase_A2_cat
IPR001937 GalP_UDPtransf1
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR036265 HIT-like_sf
IPR041373 RT_RNaseH
IPR016024 ARM-type_fold
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR008383 API5
IPR038718 SNF2-like_sf
IPR027417 P-loop_NTPase
IPR000330 SNF2_N
IPR027806 HARBI1_dom
IPR000164 Histone_H3/CENP-A
IPR002119 Histone_H2A
IPR032454 Histone_H2A_C
IPR007125 Histone_H2A/H2B/H3
IPR009072 Histone-fold
IPR018061 Retropepsins
SUPFAM
ProteinModelPortal
A0A0J7NWH7
D6WTN9
W4Y2D0
A0A085M208
W4XB88
J9KB96
+ More
W4Y5A8 A0A1X7VKM7 W4YUC1 Q9BM81 G7Y855 Q9BM79 Q9BM80 A0A0J7K7I7 W4ZBK7 C7C1Z9 A0A085MR14 A0A147BLP1 A0A085N6R2 A0A085N6P6 A0A085N5A1 A0A0C2MTE2 A0A0C2N259 X1XPJ2 X1XU40 A0A085MTV7 A0A0C2NBN0 A0A0C2MJW7 A0A0C2NDW5 X1XTW4 A0A085NRB3 K7J8H5 A0A085LN63 X1X9N5 A0A0V1F272 A0A085N387 L7LZ63 K7JCJ0 A0A147BJG9 X1XU43 K7JAG6 A0A0V0TCH2 A0A0V1FLR3 A0A085LKW1 A0A085MDV5 X1XU27 K7ISS4 A0A085LRR2 C7C1Z7 K7JBH7 A0A0V1KEE1 J9K9L2
W4Y5A8 A0A1X7VKM7 W4YUC1 Q9BM81 G7Y855 Q9BM79 Q9BM80 A0A0J7K7I7 W4ZBK7 C7C1Z9 A0A085MR14 A0A147BLP1 A0A085N6R2 A0A085N6P6 A0A085N5A1 A0A0C2MTE2 A0A0C2N259 X1XPJ2 X1XU40 A0A085MTV7 A0A0C2NBN0 A0A0C2MJW7 A0A0C2NDW5 X1XTW4 A0A085NRB3 K7J8H5 A0A085LN63 X1X9N5 A0A0V1F272 A0A085N387 L7LZ63 K7JCJ0 A0A147BJG9 X1XU43 K7JAG6 A0A0V0TCH2 A0A0V1FLR3 A0A085LKW1 A0A085MDV5 X1XU27 K7ISS4 A0A085LRR2 C7C1Z7 K7JBH7 A0A0V1KEE1 J9K9L2
PDB
4OL8
E-value=1.62127e-66,
Score=647
Ontologies
GO
Topology
Subcellular location
Chromosome
Nucleus
Nucleus
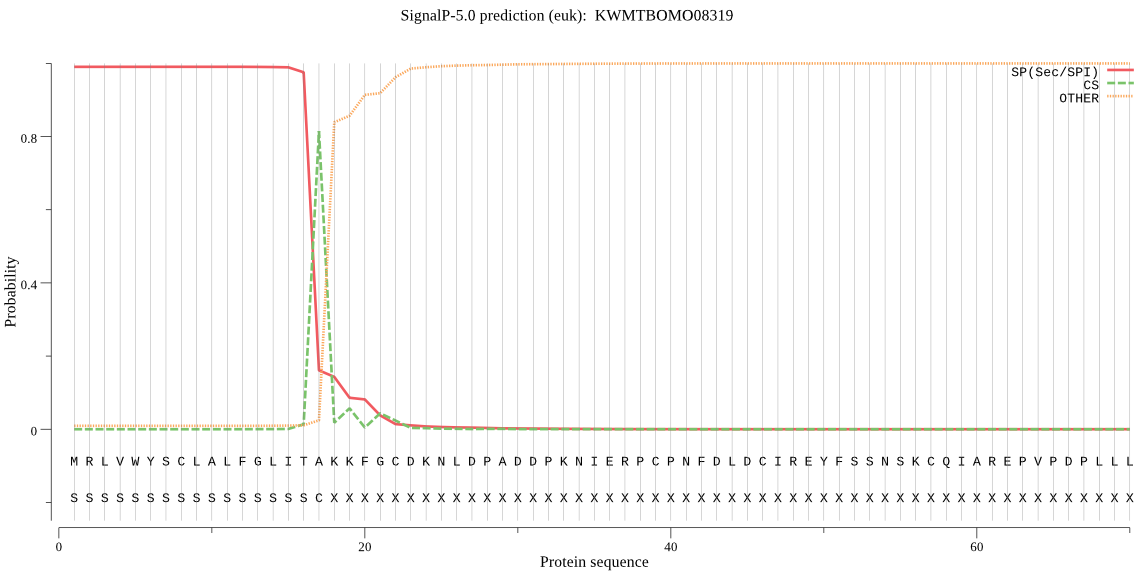
SignalP
Position: 1 - 17,
Likelihood: 0.990027
Length:
1452
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10734
Exp number, first 60 AAs:
0.09044
Total prob of N-in:
0.00573
outside
1 - 1452

Population Genetic Test Statistics
Pi
278.958756
Theta
175.970472
Tajima's D
2.287252
CLR
0
CSRT
0.925653717314134
Interpretation
Uncertain
