Gene
KWMTBOMO08318 Validated by peptides from experiments
Annotation
PREDICTED:_uncharacterized_protein_LOC105842476_[Bombyx_mori]
Full name
Fibrohexamerin
Alternative Name
25 kDa silk glycoprotein
p25
p25
Location in the cell
Nuclear Reliability : 1.535 PlasmaMembrane Reliability : 1.187
Sequence
CDS
ATGAAGTTAATGCTGTGTTTATGTCTTCTTTCGTTTTTCGGCTTGACTGTGGCGGATGAGGTAGACTGTGAATCTGATCTAGATCCGGCCGATGATCCGAGGAATATCGAAAGACCTTGTCCGAACTTCGACTTGGACTGTATTCGTAAATATTTCTCGAGTAATTCGAAATGTCAAATAACGCTGGGGCCAGTACCGGATCCGCTGCTTCTCAACAATTACAGATTAGATATAGCAAACTCTAATATTACCGCCCAATTTAACAACGTAAGCGTTCGAGGTCTCAATGGGAATATCGTTGAATTTTACATCAACAGAAAAACTGAGAAACTGGTTCTCGCAACAGAAGTGAAAAGCTTAGCATTTGATTCTCCACAGGTGGTATTTAAATACAACAGGAAAGGTAAAGAGCCCATTGTACGAGGTGACTGTGTGGATTGTGAATACGGAGCTGCAACTTTCACTGCTGTATTTCCAAGTATCTGCGATTTAGATCTGAGCAAAGCTGAAGTTTTCACCTACGTCGAAGACTCTAATCCCAGATATAAAATAGGATCACGACTATTTTCCATTTCTGATTCTGTGGCACTAAACGAGTTCCTAAAGTTTATAACGAGTATGTCGGAAAATATTCAAGAAAAGTTTATCTCGCAAGGAAGAATTTTTATGGCCAACTACATAGAATATAATATCTGTGACTTTGGATTAAATTTAGCATGTAAATAA
Protein
MKLMLCLCLLSFFGLTVADEVDCESDLDPADDPRNIERPCPNFDLDCIRKYFSSNSKCQITLGPVPDPLLLNNYRLDIANSNITAQFNNVSVRGLNGNIVEFYINRKTEKLVLATEVKSLAFDSPQVVFKYNRKGKEPIVRGDCVDCEYGAATFTAVFPSICDLDLSKAEVFTYVEDSNPRYKIGSRLFSISDSVALNEFLKFITSMSENIQEKFISQGRIFMANYIEYNICDFGLNLACK
Summary
Subunit
Silk fibroin elementary unit consists in a disulfide-linked heavy and light chain and a p25 glycoprotein in molar ratios of 6:6:1. This results in a complex of approximately 2.3 MDa.
Miscellaneous
It is unclear whether the N-terminal residue of the mature protein is Ala-17 or Gly-18.
Keywords
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Reference proteome
Secreted
Signal
Silk protein
Feature
chain Fibrohexamerin
Uniprot
Pubmed
EMBL
BABH01022228
BABH01022225
LC001863
BAS31051.1
LC001864
BAS31052.1
+ More
KZ149916 PZC77940.1 LC001865 BAS31053.1 NWSH01003760 PCG65847.1 JTDY01003497 KOB69466.1 PZC77938.1 PZC77939.1 KOB69467.1 AB001821 BAB39500.1 X04226 BP122961 BP126317 BP177743 BAAB01057839 NWSH01001474 PCG71139.1 AB001823 BAB39502.1 AB195976 BAE97692.1 BABH01038094 BABH01038095 KY792994 ARE31005.1 GQ901976 ACX50393.1 AF009827 AF009677 RSAL01000058 RVE49720.1
KZ149916 PZC77940.1 LC001865 BAS31053.1 NWSH01003760 PCG65847.1 JTDY01003497 KOB69466.1 PZC77938.1 PZC77939.1 KOB69467.1 AB001821 BAB39500.1 X04226 BP122961 BP126317 BP177743 BAAB01057839 NWSH01001474 PCG71139.1 AB001823 BAB39502.1 AB195976 BAE97692.1 BABH01038094 BABH01038095 KY792994 ARE31005.1 GQ901976 ACX50393.1 AF009827 AF009677 RSAL01000058 RVE49720.1
Proteomes
Pfam
PF07294 Fibroin_P25
Gene 3D
ProteinModelPortal
Ontologies
GO
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 18,
Likelihood: 0.999442

Length:
241
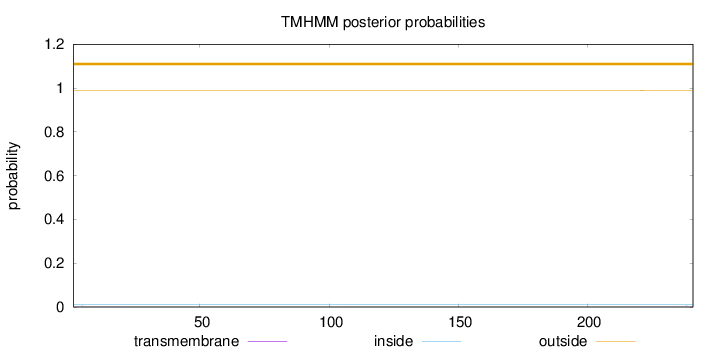
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02123
Exp number, first 60 AAs:
0.00327
Total prob of N-in:
0.01009
outside
1 - 241
Population Genetic Test Statistics
Pi
290.424774
Theta
185.816286
Tajima's D
1.653556
CLR
0.22489
CSRT
0.818959052047398
Interpretation
Uncertain
