Gene
KWMTBOMO08314
Pre Gene Modal
BGIBMGA009245
Annotation
PREDICTED:_carbonic_anhydrase-related_protein_10_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.03
Sequence
CDS
ATGCTTCAATGTTCACTTCTCATACTTACAGTCTTATGCGTCTCTATTCAGGACATCTCGGGAAGCTGGGAGGAATGGTGGACTTATGATGGTATTTCAGGTCCTGGTTTCTGGGGCCTAATAAACCCGCAATGGAGTATGTGCAATAAAGGAAGAAGACAGAGTCCAGTAAACGTGGAGCCAGATAAACTTCTCTTTGACCCGTGGCTTAGAGAAATCCAATTCGATAAACATAAGGTGAGCGGCGTACTTCAAAACACGGGCCAGTCCCTGGTCTTCCGCGTCGAGAAAGACTCGAAGCATCAGGTGAACATCAGCGGCGGGCCCCTGTCCTACCGTTACCAGTTCGAAGAGATCTACTTCCATTACGGTATGGAAGACAACAGGGGCTCAGAACATCAAATCGACCACCACACGTTTCCAGGCGAGATCCAGCTCTACGGATTCAACAAGGAACTCTATCATAACATGTCCGAAGCTCAACACAAATCCCAAGGAGTGGTTGGAATATCACTTATGGTCCAGATCGGGGAACCAACTAACAAGGAACTGCGACTGATAACCAGCGCATTCAACAAAGTTACTTACCGAGGCAGCTCCTACCCAATCAAGCATCTGCCTCTGAGTTCTTTGTTGCCGAACACCCAGCAGTACCTTACTTATGAAGGCTCGACTACGCACCCGGGTTGCTGGGAGACAGCCGTTTGGATAATATTCAACAAACCCATTTATATCTCTAAGCAAGAGATGTATGCACTCCGAAGACTGATGCAAGGGTCGCAGTTGACTCCAAAAGCTCCACTGGGTAACAACGCTCGTCCCGTCCAGCCTCTCCACCATCGCACTGTAAGAACGAACATCAACTTCAATAAACAAGGAGTACAGGTCTCGAGCAACTGTCCCGATATGTACAGGAATATGCATTACACAGCAACTCAGTGGCCGCGGGATCACAACATTCGCTACAGAAACATAGACGATCTCGCTCTACCGATAAACTAA
Protein
MLQCSLLILTVLCVSIQDISGSWEEWWTYDGISGPGFWGLINPQWSMCNKGRRQSPVNVEPDKLLFDPWLREIQFDKHKVSGVLQNTGQSLVFRVEKDSKHQVNISGGPLSYRYQFEEIYFHYGMEDNRGSEHQIDHHTFPGEIQLYGFNKELYHNMSEAQHKSQGVVGISLMVQIGEPTNKELRLITSAFNKVTYRGSSYPIKHLPLSSLLPNTQQYLTYEGSTTHPGCWETAVWIIFNKPIYISKQEMYALRRLMQGSQLTPKAPLGNNARPVQPLHHRTVRTNINFNKQGVQVSSNCPDMYRNMHYTATQWPRDHNIRYRNIDDLALPIN
Summary
Similarity
Belongs to the alpha-carbonic anhydrase family.
Uniprot
A0A2W1BXI8
A0A194PZN9
A0A437BRH7
S4P9P5
A0A212EPP1
A0A194RC42
+ More
H9JI95 A0A1Q3FV96 A0A2C9H8A9 Q7PRN8 A0A1S4GAD3 C0KJQ8 A0A453YJZ5 Q16ZY1 A0A182QJE7 A0A0M4FB19 A0A0Q9WW22 B4MAF5 A0A3B0JX60 B5DMT7 B4GVV9 B4L6B3 B3MRB2 B4Q0S4 B4IL36 B3NTT1 Q9W3P7 A0A1W4VK37 W8C9C5 A0A0L0BRC4 A0A1J1IUX8 A0A1I8MMK0 B4JK33 B4R682 A0A182Y7K5 A0A182J6K1 A0A1B6FQG3 A0A1B6C938 A0A1B6GY94 A0A1B6HEQ7 A0A1B6J6B5 A0A1B6MGD2 A0A0A9XLS0 A0A336LGW7 D2A110 A0A1B6C4Y5 A0A224XUC8 A0A1B6FC64 A0A1B6HNG7 A0A1B6JTD6 A0A1B6HXS6 A0A1B6MJC1 A0A1A9Z5T6 A0A182MZR5 A0A0C9RQ62 A0A182PTG1 A0A3L8DWM0 B0XKW2 K7J6C4 A0A2M4CK18 A0A336MXL1 A0A1W4WSQ2 A0A2J7RQD8 A0A182R3H1 A0A1I8P8P1 A0A1A9XXS2 A0A088A1J3 A0A182W4B9 A0A026WAC8 E0VKF0 A0A232FBF2 A0A1W4X3M2 A0A0P5QRT2 A0A162C736 A0A0P5GGG6 A0A0P5QIK4 A0A0P6APQ9 A0A0N8E6G9 A0A0P5K439 E9FWT4 A0A2A4JPU0 A0A0N8A793 D2A0V8 E2AWI0 A0A2H8TCM9 J9JLX4 A0A1B6DUA5 A0A0P4XIA3 A0A0L7QNK1 A0A1Y1N5R7 A0A212EYD6 A0A194RLP6 A0A158P0L6 A0A2S2NYK4 A0A087ZUM2 K7ISF5 A0A0P5LGV2 A0A1S3D1H3 A0A232EJ82
H9JI95 A0A1Q3FV96 A0A2C9H8A9 Q7PRN8 A0A1S4GAD3 C0KJQ8 A0A453YJZ5 Q16ZY1 A0A182QJE7 A0A0M4FB19 A0A0Q9WW22 B4MAF5 A0A3B0JX60 B5DMT7 B4GVV9 B4L6B3 B3MRB2 B4Q0S4 B4IL36 B3NTT1 Q9W3P7 A0A1W4VK37 W8C9C5 A0A0L0BRC4 A0A1J1IUX8 A0A1I8MMK0 B4JK33 B4R682 A0A182Y7K5 A0A182J6K1 A0A1B6FQG3 A0A1B6C938 A0A1B6GY94 A0A1B6HEQ7 A0A1B6J6B5 A0A1B6MGD2 A0A0A9XLS0 A0A336LGW7 D2A110 A0A1B6C4Y5 A0A224XUC8 A0A1B6FC64 A0A1B6HNG7 A0A1B6JTD6 A0A1B6HXS6 A0A1B6MJC1 A0A1A9Z5T6 A0A182MZR5 A0A0C9RQ62 A0A182PTG1 A0A3L8DWM0 B0XKW2 K7J6C4 A0A2M4CK18 A0A336MXL1 A0A1W4WSQ2 A0A2J7RQD8 A0A182R3H1 A0A1I8P8P1 A0A1A9XXS2 A0A088A1J3 A0A182W4B9 A0A026WAC8 E0VKF0 A0A232FBF2 A0A1W4X3M2 A0A0P5QRT2 A0A162C736 A0A0P5GGG6 A0A0P5QIK4 A0A0P6APQ9 A0A0N8E6G9 A0A0P5K439 E9FWT4 A0A2A4JPU0 A0A0N8A793 D2A0V8 E2AWI0 A0A2H8TCM9 J9JLX4 A0A1B6DUA5 A0A0P4XIA3 A0A0L7QNK1 A0A1Y1N5R7 A0A212EYD6 A0A194RLP6 A0A158P0L6 A0A2S2NYK4 A0A087ZUM2 K7ISF5 A0A0P5LGV2 A0A1S3D1H3 A0A232EJ82
Pubmed
28756777
26354079
23622113
22118469
19121390
12364791
+ More
17981859 17510324 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 26108605 25315136 25244985 25401762 18362917 19820115 30249741 20075255 24508170 20566863 28648823 21292972 20798317 28004739 21347285
17981859 17510324 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 26108605 25315136 25244985 25401762 18362917 19820115 30249741 20075255 24508170 20566863 28648823 21292972 20798317 28004739 21347285
EMBL
KZ149916
PZC77937.1
KQ459585
KPI98209.1
RSAL01000016
RVE53054.1
+ More
GAIX01005371 JAA87189.1 AGBW02013437 OWR43439.1 KQ460627 KPJ13476.1 BABH01022223 BABH01022224 GFDL01003570 JAV31475.1 AAAB01008847 EAA06900.6 FJ612599 ACM89452.1 APCN01001035 APCN01001036 APCN01001037 CH477481 EAT40222.1 AXCN02001216 CP012528 ALC49720.1 CH964239 KRF99656.1 CH940655 EDW66214.2 OUUW01000003 SPP78339.1 CH379064 EDY73119.1 CH479193 EDW26804.1 CH933812 EDW05909.2 CH902622 EDV34317.1 CM000162 EDX02345.2 CH480865 EDW53653.1 CH954180 EDV47494.1 AE014298 BT024405 AAF46274.4 ABC86467.1 AHN59457.1 GAMC01002279 JAC04277.1 JRES01001480 KNC22587.1 CVRI01000057 CRL02345.1 CH916370 EDV99935.1 CM000366 EDX17359.1 GECZ01017390 JAS52379.1 GEDC01027548 JAS09750.1 GECZ01002380 JAS67389.1 GECU01034627 JAS73079.1 GECU01012990 JAS94716.1 GEBQ01005028 JAT34949.1 GBHO01023023 JAG20581.1 UFQS01003758 UFQT01003758 SSX15913.1 SSX35255.1 KQ971338 EFA02589.2 GEDC01028732 JAS08566.1 GFTR01004875 JAW11551.1 GECZ01021977 JAS47792.1 GECU01031515 JAS76191.1 GECU01005251 JAT02456.1 GECU01028220 JAS79486.1 GEBQ01004028 JAT35949.1 GBYB01009331 JAG79098.1 QOIP01000003 RLU24633.1 DS234059 EDS32747.1 GGFL01001524 MBW65702.1 UFQS01002637 UFQT01002637 SSX14419.1 SSX33829.1 NEVH01001340 PNF43045.1 KK107295 EZA53047.1 DS235243 EEB13866.1 NNAY01000504 OXU27952.1 GDIQ01117353 GDIP01047815 JAL34373.1 JAM55900.1 LRGB01001361 KZS12302.1 GDIQ01241308 JAK10417.1 GDIQ01113647 JAL38079.1 GDIP01028000 JAM75715.1 GDIQ01057116 JAN37621.1 GDIQ01194968 JAK56757.1 GL732526 EFX88377.1 NWSH01000817 PCG74067.1 GDIP01175593 JAJ47809.1 EFA01627.2 GL443297 EFN62208.1 GFXV01000066 MBW11871.1 ABLF02023847 ABLF02026736 ABLF02026737 ABLF02026739 ABLF02026740 GEDC01008055 JAS29243.1 GDIP01241435 JAI81966.1 KQ414851 KOC60205.1 GEZM01014016 JAV92220.1 AGBW02011532 OWR46506.1 KQ460205 KPJ16906.1 ADTU01005290 ADTU01005291 GGMR01009419 MBY22038.1 GDIQ01194969 JAK56756.1 NNAY01004105 OXU18361.1
GAIX01005371 JAA87189.1 AGBW02013437 OWR43439.1 KQ460627 KPJ13476.1 BABH01022223 BABH01022224 GFDL01003570 JAV31475.1 AAAB01008847 EAA06900.6 FJ612599 ACM89452.1 APCN01001035 APCN01001036 APCN01001037 CH477481 EAT40222.1 AXCN02001216 CP012528 ALC49720.1 CH964239 KRF99656.1 CH940655 EDW66214.2 OUUW01000003 SPP78339.1 CH379064 EDY73119.1 CH479193 EDW26804.1 CH933812 EDW05909.2 CH902622 EDV34317.1 CM000162 EDX02345.2 CH480865 EDW53653.1 CH954180 EDV47494.1 AE014298 BT024405 AAF46274.4 ABC86467.1 AHN59457.1 GAMC01002279 JAC04277.1 JRES01001480 KNC22587.1 CVRI01000057 CRL02345.1 CH916370 EDV99935.1 CM000366 EDX17359.1 GECZ01017390 JAS52379.1 GEDC01027548 JAS09750.1 GECZ01002380 JAS67389.1 GECU01034627 JAS73079.1 GECU01012990 JAS94716.1 GEBQ01005028 JAT34949.1 GBHO01023023 JAG20581.1 UFQS01003758 UFQT01003758 SSX15913.1 SSX35255.1 KQ971338 EFA02589.2 GEDC01028732 JAS08566.1 GFTR01004875 JAW11551.1 GECZ01021977 JAS47792.1 GECU01031515 JAS76191.1 GECU01005251 JAT02456.1 GECU01028220 JAS79486.1 GEBQ01004028 JAT35949.1 GBYB01009331 JAG79098.1 QOIP01000003 RLU24633.1 DS234059 EDS32747.1 GGFL01001524 MBW65702.1 UFQS01002637 UFQT01002637 SSX14419.1 SSX33829.1 NEVH01001340 PNF43045.1 KK107295 EZA53047.1 DS235243 EEB13866.1 NNAY01000504 OXU27952.1 GDIQ01117353 GDIP01047815 JAL34373.1 JAM55900.1 LRGB01001361 KZS12302.1 GDIQ01241308 JAK10417.1 GDIQ01113647 JAL38079.1 GDIP01028000 JAM75715.1 GDIQ01057116 JAN37621.1 GDIQ01194968 JAK56757.1 GL732526 EFX88377.1 NWSH01000817 PCG74067.1 GDIP01175593 JAJ47809.1 EFA01627.2 GL443297 EFN62208.1 GFXV01000066 MBW11871.1 ABLF02023847 ABLF02026736 ABLF02026737 ABLF02026739 ABLF02026740 GEDC01008055 JAS29243.1 GDIP01241435 JAI81966.1 KQ414851 KOC60205.1 GEZM01014016 JAV92220.1 AGBW02011532 OWR46506.1 KQ460205 KPJ16906.1 ADTU01005290 ADTU01005291 GGMR01009419 MBY22038.1 GDIQ01194969 JAK56756.1 NNAY01004105 OXU18361.1
Proteomes
UP000053268
UP000283053
UP000007151
UP000053240
UP000005204
UP000076407
+ More
UP000007062 UP000075840 UP000008820 UP000075886 UP000092553 UP000007798 UP000008792 UP000268350 UP000001819 UP000008744 UP000009192 UP000007801 UP000002282 UP000001292 UP000008711 UP000000803 UP000192221 UP000037069 UP000183832 UP000095301 UP000001070 UP000000304 UP000076408 UP000075880 UP000007266 UP000092445 UP000075884 UP000075885 UP000279307 UP000002320 UP000002358 UP000192223 UP000235965 UP000075900 UP000095300 UP000092443 UP000005203 UP000075920 UP000053097 UP000009046 UP000215335 UP000076858 UP000000305 UP000218220 UP000000311 UP000007819 UP000053825 UP000005205 UP000079169
UP000007062 UP000075840 UP000008820 UP000075886 UP000092553 UP000007798 UP000008792 UP000268350 UP000001819 UP000008744 UP000009192 UP000007801 UP000002282 UP000001292 UP000008711 UP000000803 UP000192221 UP000037069 UP000183832 UP000095301 UP000001070 UP000000304 UP000076408 UP000075880 UP000007266 UP000092445 UP000075884 UP000075885 UP000279307 UP000002320 UP000002358 UP000192223 UP000235965 UP000075900 UP000095300 UP000092443 UP000005203 UP000075920 UP000053097 UP000009046 UP000215335 UP000076858 UP000000305 UP000218220 UP000000311 UP000007819 UP000053825 UP000005205 UP000079169
Pfam
PF00194 Carb_anhydrase
Interpro
SUPFAM
SSF51069
SSF51069
Gene 3D
ProteinModelPortal
A0A2W1BXI8
A0A194PZN9
A0A437BRH7
S4P9P5
A0A212EPP1
A0A194RC42
+ More
H9JI95 A0A1Q3FV96 A0A2C9H8A9 Q7PRN8 A0A1S4GAD3 C0KJQ8 A0A453YJZ5 Q16ZY1 A0A182QJE7 A0A0M4FB19 A0A0Q9WW22 B4MAF5 A0A3B0JX60 B5DMT7 B4GVV9 B4L6B3 B3MRB2 B4Q0S4 B4IL36 B3NTT1 Q9W3P7 A0A1W4VK37 W8C9C5 A0A0L0BRC4 A0A1J1IUX8 A0A1I8MMK0 B4JK33 B4R682 A0A182Y7K5 A0A182J6K1 A0A1B6FQG3 A0A1B6C938 A0A1B6GY94 A0A1B6HEQ7 A0A1B6J6B5 A0A1B6MGD2 A0A0A9XLS0 A0A336LGW7 D2A110 A0A1B6C4Y5 A0A224XUC8 A0A1B6FC64 A0A1B6HNG7 A0A1B6JTD6 A0A1B6HXS6 A0A1B6MJC1 A0A1A9Z5T6 A0A182MZR5 A0A0C9RQ62 A0A182PTG1 A0A3L8DWM0 B0XKW2 K7J6C4 A0A2M4CK18 A0A336MXL1 A0A1W4WSQ2 A0A2J7RQD8 A0A182R3H1 A0A1I8P8P1 A0A1A9XXS2 A0A088A1J3 A0A182W4B9 A0A026WAC8 E0VKF0 A0A232FBF2 A0A1W4X3M2 A0A0P5QRT2 A0A162C736 A0A0P5GGG6 A0A0P5QIK4 A0A0P6APQ9 A0A0N8E6G9 A0A0P5K439 E9FWT4 A0A2A4JPU0 A0A0N8A793 D2A0V8 E2AWI0 A0A2H8TCM9 J9JLX4 A0A1B6DUA5 A0A0P4XIA3 A0A0L7QNK1 A0A1Y1N5R7 A0A212EYD6 A0A194RLP6 A0A158P0L6 A0A2S2NYK4 A0A087ZUM2 K7ISF5 A0A0P5LGV2 A0A1S3D1H3 A0A232EJ82
H9JI95 A0A1Q3FV96 A0A2C9H8A9 Q7PRN8 A0A1S4GAD3 C0KJQ8 A0A453YJZ5 Q16ZY1 A0A182QJE7 A0A0M4FB19 A0A0Q9WW22 B4MAF5 A0A3B0JX60 B5DMT7 B4GVV9 B4L6B3 B3MRB2 B4Q0S4 B4IL36 B3NTT1 Q9W3P7 A0A1W4VK37 W8C9C5 A0A0L0BRC4 A0A1J1IUX8 A0A1I8MMK0 B4JK33 B4R682 A0A182Y7K5 A0A182J6K1 A0A1B6FQG3 A0A1B6C938 A0A1B6GY94 A0A1B6HEQ7 A0A1B6J6B5 A0A1B6MGD2 A0A0A9XLS0 A0A336LGW7 D2A110 A0A1B6C4Y5 A0A224XUC8 A0A1B6FC64 A0A1B6HNG7 A0A1B6JTD6 A0A1B6HXS6 A0A1B6MJC1 A0A1A9Z5T6 A0A182MZR5 A0A0C9RQ62 A0A182PTG1 A0A3L8DWM0 B0XKW2 K7J6C4 A0A2M4CK18 A0A336MXL1 A0A1W4WSQ2 A0A2J7RQD8 A0A182R3H1 A0A1I8P8P1 A0A1A9XXS2 A0A088A1J3 A0A182W4B9 A0A026WAC8 E0VKF0 A0A232FBF2 A0A1W4X3M2 A0A0P5QRT2 A0A162C736 A0A0P5GGG6 A0A0P5QIK4 A0A0P6APQ9 A0A0N8E6G9 A0A0P5K439 E9FWT4 A0A2A4JPU0 A0A0N8A793 D2A0V8 E2AWI0 A0A2H8TCM9 J9JLX4 A0A1B6DUA5 A0A0P4XIA3 A0A0L7QNK1 A0A1Y1N5R7 A0A212EYD6 A0A194RLP6 A0A158P0L6 A0A2S2NYK4 A0A087ZUM2 K7ISF5 A0A0P5LGV2 A0A1S3D1H3 A0A232EJ82
PDB
1Z97
E-value=4.88276e-33,
Score=352
Ontologies
PANTHER
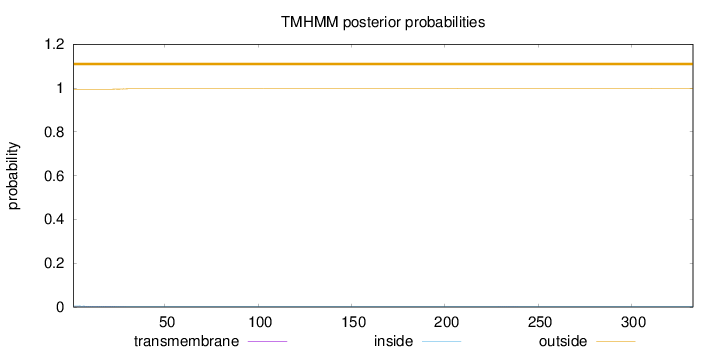
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.982204

Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03825
Exp number, first 60 AAs:
0.03571
Total prob of N-in:
0.00511
outside
1 - 333
Population Genetic Test Statistics
Pi
178.349793
Theta
168.020081
Tajima's D
-1.525299
CLR
138.067332
CSRT
0.0585970701464927
Interpretation
Uncertain
