Gene
KWMTBOMO08313
Pre Gene Modal
BGIBMGA009247
Annotation
PREDICTED:_alpha-(1?3)-fucosyltransferase_C-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.204 Mitochondrial Reliability : 1.427
Sequence
CDS
ATGAGCTATCAATGGCCTGACCGAATGGAAGCCATCATTGATCCCGTGTTCAGAGAGGAGCTGAACTCAAAAGCCAAAGGAGTCCTTACCTTTCTTGACAAATGCCAGAGTAGAAGCAAAAGAGAGGTGTTCTTGAAGAACCTTCAGAATGAATTATCAAAATATAACCTGACTGTGGATATTTTTGGTAAATGTGGTCGAGCCTGCAGCCCGAGACGTAAGTCAATGAAACCATGTTTCTGGAAATTGAGGAAGGATTATTTCTTCTATTTGGCAATGGAAGATTCTTTCGCTAAAGATTTTATGACCGATAGCGTACTTTATGGGTTAAAGTATAATTCAGTGCCTATCGTTTACGGGAAGGCTGATTATGACAAATTTCTACCACCGAATTCATACATTAACGCTATGGAACTTGGAGAAGAGAAATTAGCAAAAACAATAAACACGATTATGAGAAACATGCAAATTTACTACGAGTTCTTCGCGTGGAGGAACCATTACGCCGTCAGGGAAATGCAGGTCCTTGACCCTTGCTCTTTGTGCGATGCTTTAAATAAACCCGAGTGGTTGGAGTACCGGTGGAGTCACAGTCAATTCAGGAATTGGTGGAACCCTGAATACGAGGACCGATGCCCGTTTTATAGTAAATATTTAACGCTTTAA
Protein
MSYQWPDRMEAIIDPVFREELNSKAKGVLTFLDKCQSRSKREVFLKNLQNELSKYNLTVDIFGKCGRACSPRRKSMKPCFWKLRKDYFFYLAMEDSFAKDFMTDSVLYGLKYNSVPIVYGKADYDKFLPPNSYINAMELGEEKLAKTINTIMRNMQIYYEFFAWRNHYAVREMQVLDPCSLCDALNKPEWLEYRWSHSQFRNWWNPEYEDRCPFYSKYLTL
Summary
Similarity
Belongs to the glycosyltransferase 10 family.
Uniprot
H9JI97
A0A3S2PJA5
A0A212FHM7
A0A2H1V6K5
A0A212F4A2
A0A3S2LG40
+ More
A0A0L7KT82 A0A194R7I6 A0A2W1BS87 A0A194QTG1 A0A3S2NPV0 A0A212FDW4 A0A0L7LD09 A0A194R6N8 A0A194R740 A0A194QV22 H9JI93 A0A212EZY5 A0A2H1V9M8 A0A212FE36 A0A2H1V6R4 A0A194PTI0 A0A194Q472 A0A2A4J2A2 A0A2A4J733 A0A2H1WPY7 A0A0L7KKB0 A0A3S2LHN9 A0A0L7K4N4 A0A212EPM2 A0A2H1VN94 A0A2H1X3C4 A0A2W1C1F3 A0A2A4J5A9 A0A2A4J5I4 A0A2H1VKP0 A0A3S2NZG0 A0A3S2LQB6 A0A3S2M564 A0A3S2M779 A0A0L7LBT7 A0A2A4J0X4 A0A2W1BZ24 A0A3S2NK67 A0A2W1BWB4 A0A2H1WHJ5 A0A2A4JYW9 A0A437BMJ2 A0A2H1VCS8 A0A3S2PHJ4 A0A437BN03 A0A2A4J738 A0A2A4J2N7 A0A212F5Z8 A0A2W1BGR4 A0A154PNF0 U5EV02 A0A194PY00 A0A2A4J121 A0A2H1VDQ5 E9HNE7 E9HNE2 A0A0P5JKQ6 A0A3S2NI47 E9IYL6 A0A0P5IGH3 A0A194R892 A0A0P5NF53 A0A0P5MPQ9 A0A310SMV6 A0A0P6G0S5 A0A0P5H7W2 A0A0P5KZU0 A0A0P5PYI1 A0A0P6GR73 A0A0P5T191 A0A0P6GWX6 A0A0P5P0I8 A0A0P6DCY4 A0A1D2MJA3 A0A1W4WDM3 A0A0P5NT39 A0A0P6HBD3 A0A1W4WCP4 E2AUH5 A0A0P5VXP3 A0A0P5WXM8 A0A0P5JXK7 A0A0P5CWM3 A0A2H1VRA0 B4L403 A0A0P6CTY6 A0A0P5SV44 E9FXZ7 A0A0P5BKE4 E2BU28 A0A0P5HM40 A0A0P5N0G2 A0A0P5VKZ5 D6WT64
A0A0L7KT82 A0A194R7I6 A0A2W1BS87 A0A194QTG1 A0A3S2NPV0 A0A212FDW4 A0A0L7LD09 A0A194R6N8 A0A194R740 A0A194QV22 H9JI93 A0A212EZY5 A0A2H1V9M8 A0A212FE36 A0A2H1V6R4 A0A194PTI0 A0A194Q472 A0A2A4J2A2 A0A2A4J733 A0A2H1WPY7 A0A0L7KKB0 A0A3S2LHN9 A0A0L7K4N4 A0A212EPM2 A0A2H1VN94 A0A2H1X3C4 A0A2W1C1F3 A0A2A4J5A9 A0A2A4J5I4 A0A2H1VKP0 A0A3S2NZG0 A0A3S2LQB6 A0A3S2M564 A0A3S2M779 A0A0L7LBT7 A0A2A4J0X4 A0A2W1BZ24 A0A3S2NK67 A0A2W1BWB4 A0A2H1WHJ5 A0A2A4JYW9 A0A437BMJ2 A0A2H1VCS8 A0A3S2PHJ4 A0A437BN03 A0A2A4J738 A0A2A4J2N7 A0A212F5Z8 A0A2W1BGR4 A0A154PNF0 U5EV02 A0A194PY00 A0A2A4J121 A0A2H1VDQ5 E9HNE7 E9HNE2 A0A0P5JKQ6 A0A3S2NI47 E9IYL6 A0A0P5IGH3 A0A194R892 A0A0P5NF53 A0A0P5MPQ9 A0A310SMV6 A0A0P6G0S5 A0A0P5H7W2 A0A0P5KZU0 A0A0P5PYI1 A0A0P6GR73 A0A0P5T191 A0A0P6GWX6 A0A0P5P0I8 A0A0P6DCY4 A0A1D2MJA3 A0A1W4WDM3 A0A0P5NT39 A0A0P6HBD3 A0A1W4WCP4 E2AUH5 A0A0P5VXP3 A0A0P5WXM8 A0A0P5JXK7 A0A0P5CWM3 A0A2H1VRA0 B4L403 A0A0P6CTY6 A0A0P5SV44 E9FXZ7 A0A0P5BKE4 E2BU28 A0A0P5HM40 A0A0P5N0G2 A0A0P5VKZ5 D6WT64
Pubmed
EMBL
BABH01022212
RSAL01000016
RVE53050.1
AGBW02008491
OWR53234.1
ODYU01000952
+ More
SOQ36485.1 AGBW02010417 OWR48543.1 RVE53056.1 JTDY01005919 KOB66482.1 KQ460627 KPJ13479.1 KZ149916 PZC77942.1 KQ461137 KPJ08813.1 RSAL01000164 RVE45329.1 AGBW02008995 OWR51946.1 JTDY01001718 KOB73086.1 KPJ13478.1 KPJ13477.1 KQ461108 KPJ09317.1 BABH01022242 AGBW02011221 OWR47017.1 ODYU01001405 SOQ37538.1 OWR51947.1 SOQ36489.1 KQ459594 KPI96064.1 KQ459585 KPI98210.1 NWSH01003936 PCG65654.1 NWSH01002952 PCG67232.1 ODYU01010180 SOQ55129.1 JTDY01009552 KOB63349.1 RSAL01000116 RVE46859.1 JTDY01010816 KOB57987.1 AGBW02013437 OWR43438.1 ODYU01003471 SOQ42305.1 ODYU01013118 SOQ59805.1 PZC77933.1 NWSH01003242 PCG66704.1 PCG67231.1 ODYU01003079 SOQ41387.1 RVE53053.1 RSAL01000031 RVE51640.1 RVE51643.1 RVE53052.1 JTDY01001793 KOB72859.1 NWSH01004476 PCG65062.1 PZC77936.1 RVE53051.1 PZC77935.1 ODYU01008712 SOQ52539.1 NWSH01000372 PCG76878.1 RVE51644.1 ODYU01001860 SOQ38649.1 RVE51641.1 RVE51638.1 PCG67233.1 PCG65653.1 AGBW02010111 OWR49153.1 KZ150145 PZC72905.1 KQ434977 KZC12838.1 GANO01003632 JAB56239.1 KPI98207.1 NWSH01004327 PCG65224.1 ODYU01002011 SOQ38990.1 GL732695 EFX66744.1 EFX66743.1 GDIQ01198740 JAK52985.1 RVE51642.1 GL767018 EFZ14350.1 GDIQ01215036 JAK36689.1 KPJ13475.1 GDIQ01153764 JAK97961.1 GDIQ01153765 JAK97960.1 KQ761844 OAD56586.1 GDIQ01040217 JAN54520.1 GDIQ01230053 JAK21672.1 GDIQ01178058 JAK73667.1 GDIQ01122417 JAL29309.1 GDIQ01029575 JAN65162.1 GDIQ01097525 JAL54201.1 GDIQ01040218 JAN54519.1 GDIQ01156313 JAK95412.1 GDIQ01080709 JAN14028.1 LJIJ01001068 ODM93136.1 GDIQ01144871 JAL06855.1 GDIQ01022109 JAN72628.1 GL442820 EFN62934.1 GDIP01093781 JAM09934.1 GDIP01080086 JAM23629.1 GDIQ01193841 JAK57884.1 GDIP01164396 JAJ59006.1 ODYU01003968 SOQ43373.1 CH933810 EDW07281.1 GDIP01003226 JAN00490.1 GDIP01134841 JAL68873.1 GL732526 EFX88202.1 GDIP01198295 JAJ25107.1 GL450575 EFN80802.1 GDIQ01248391 JAK03334.1 GDIQ01150964 JAL00762.1 GDIP01113298 LRGB01001229 JAL90416.1 KZS13314.1 KQ971354 EFA05857.2
SOQ36485.1 AGBW02010417 OWR48543.1 RVE53056.1 JTDY01005919 KOB66482.1 KQ460627 KPJ13479.1 KZ149916 PZC77942.1 KQ461137 KPJ08813.1 RSAL01000164 RVE45329.1 AGBW02008995 OWR51946.1 JTDY01001718 KOB73086.1 KPJ13478.1 KPJ13477.1 KQ461108 KPJ09317.1 BABH01022242 AGBW02011221 OWR47017.1 ODYU01001405 SOQ37538.1 OWR51947.1 SOQ36489.1 KQ459594 KPI96064.1 KQ459585 KPI98210.1 NWSH01003936 PCG65654.1 NWSH01002952 PCG67232.1 ODYU01010180 SOQ55129.1 JTDY01009552 KOB63349.1 RSAL01000116 RVE46859.1 JTDY01010816 KOB57987.1 AGBW02013437 OWR43438.1 ODYU01003471 SOQ42305.1 ODYU01013118 SOQ59805.1 PZC77933.1 NWSH01003242 PCG66704.1 PCG67231.1 ODYU01003079 SOQ41387.1 RVE53053.1 RSAL01000031 RVE51640.1 RVE51643.1 RVE53052.1 JTDY01001793 KOB72859.1 NWSH01004476 PCG65062.1 PZC77936.1 RVE53051.1 PZC77935.1 ODYU01008712 SOQ52539.1 NWSH01000372 PCG76878.1 RVE51644.1 ODYU01001860 SOQ38649.1 RVE51641.1 RVE51638.1 PCG67233.1 PCG65653.1 AGBW02010111 OWR49153.1 KZ150145 PZC72905.1 KQ434977 KZC12838.1 GANO01003632 JAB56239.1 KPI98207.1 NWSH01004327 PCG65224.1 ODYU01002011 SOQ38990.1 GL732695 EFX66744.1 EFX66743.1 GDIQ01198740 JAK52985.1 RVE51642.1 GL767018 EFZ14350.1 GDIQ01215036 JAK36689.1 KPJ13475.1 GDIQ01153764 JAK97961.1 GDIQ01153765 JAK97960.1 KQ761844 OAD56586.1 GDIQ01040217 JAN54520.1 GDIQ01230053 JAK21672.1 GDIQ01178058 JAK73667.1 GDIQ01122417 JAL29309.1 GDIQ01029575 JAN65162.1 GDIQ01097525 JAL54201.1 GDIQ01040218 JAN54519.1 GDIQ01156313 JAK95412.1 GDIQ01080709 JAN14028.1 LJIJ01001068 ODM93136.1 GDIQ01144871 JAL06855.1 GDIQ01022109 JAN72628.1 GL442820 EFN62934.1 GDIP01093781 JAM09934.1 GDIP01080086 JAM23629.1 GDIQ01193841 JAK57884.1 GDIP01164396 JAJ59006.1 ODYU01003968 SOQ43373.1 CH933810 EDW07281.1 GDIP01003226 JAN00490.1 GDIP01134841 JAL68873.1 GL732526 EFX88202.1 GDIP01198295 JAJ25107.1 GL450575 EFN80802.1 GDIQ01248391 JAK03334.1 GDIQ01150964 JAL00762.1 GDIP01113298 LRGB01001229 JAL90416.1 KZS13314.1 KQ971354 EFA05857.2
Proteomes
Gene 3D
ProteinModelPortal
H9JI97
A0A3S2PJA5
A0A212FHM7
A0A2H1V6K5
A0A212F4A2
A0A3S2LG40
+ More
A0A0L7KT82 A0A194R7I6 A0A2W1BS87 A0A194QTG1 A0A3S2NPV0 A0A212FDW4 A0A0L7LD09 A0A194R6N8 A0A194R740 A0A194QV22 H9JI93 A0A212EZY5 A0A2H1V9M8 A0A212FE36 A0A2H1V6R4 A0A194PTI0 A0A194Q472 A0A2A4J2A2 A0A2A4J733 A0A2H1WPY7 A0A0L7KKB0 A0A3S2LHN9 A0A0L7K4N4 A0A212EPM2 A0A2H1VN94 A0A2H1X3C4 A0A2W1C1F3 A0A2A4J5A9 A0A2A4J5I4 A0A2H1VKP0 A0A3S2NZG0 A0A3S2LQB6 A0A3S2M564 A0A3S2M779 A0A0L7LBT7 A0A2A4J0X4 A0A2W1BZ24 A0A3S2NK67 A0A2W1BWB4 A0A2H1WHJ5 A0A2A4JYW9 A0A437BMJ2 A0A2H1VCS8 A0A3S2PHJ4 A0A437BN03 A0A2A4J738 A0A2A4J2N7 A0A212F5Z8 A0A2W1BGR4 A0A154PNF0 U5EV02 A0A194PY00 A0A2A4J121 A0A2H1VDQ5 E9HNE7 E9HNE2 A0A0P5JKQ6 A0A3S2NI47 E9IYL6 A0A0P5IGH3 A0A194R892 A0A0P5NF53 A0A0P5MPQ9 A0A310SMV6 A0A0P6G0S5 A0A0P5H7W2 A0A0P5KZU0 A0A0P5PYI1 A0A0P6GR73 A0A0P5T191 A0A0P6GWX6 A0A0P5P0I8 A0A0P6DCY4 A0A1D2MJA3 A0A1W4WDM3 A0A0P5NT39 A0A0P6HBD3 A0A1W4WCP4 E2AUH5 A0A0P5VXP3 A0A0P5WXM8 A0A0P5JXK7 A0A0P5CWM3 A0A2H1VRA0 B4L403 A0A0P6CTY6 A0A0P5SV44 E9FXZ7 A0A0P5BKE4 E2BU28 A0A0P5HM40 A0A0P5N0G2 A0A0P5VKZ5 D6WT64
A0A0L7KT82 A0A194R7I6 A0A2W1BS87 A0A194QTG1 A0A3S2NPV0 A0A212FDW4 A0A0L7LD09 A0A194R6N8 A0A194R740 A0A194QV22 H9JI93 A0A212EZY5 A0A2H1V9M8 A0A212FE36 A0A2H1V6R4 A0A194PTI0 A0A194Q472 A0A2A4J2A2 A0A2A4J733 A0A2H1WPY7 A0A0L7KKB0 A0A3S2LHN9 A0A0L7K4N4 A0A212EPM2 A0A2H1VN94 A0A2H1X3C4 A0A2W1C1F3 A0A2A4J5A9 A0A2A4J5I4 A0A2H1VKP0 A0A3S2NZG0 A0A3S2LQB6 A0A3S2M564 A0A3S2M779 A0A0L7LBT7 A0A2A4J0X4 A0A2W1BZ24 A0A3S2NK67 A0A2W1BWB4 A0A2H1WHJ5 A0A2A4JYW9 A0A437BMJ2 A0A2H1VCS8 A0A3S2PHJ4 A0A437BN03 A0A2A4J738 A0A2A4J2N7 A0A212F5Z8 A0A2W1BGR4 A0A154PNF0 U5EV02 A0A194PY00 A0A2A4J121 A0A2H1VDQ5 E9HNE7 E9HNE2 A0A0P5JKQ6 A0A3S2NI47 E9IYL6 A0A0P5IGH3 A0A194R892 A0A0P5NF53 A0A0P5MPQ9 A0A310SMV6 A0A0P6G0S5 A0A0P5H7W2 A0A0P5KZU0 A0A0P5PYI1 A0A0P6GR73 A0A0P5T191 A0A0P6GWX6 A0A0P5P0I8 A0A0P6DCY4 A0A1D2MJA3 A0A1W4WDM3 A0A0P5NT39 A0A0P6HBD3 A0A1W4WCP4 E2AUH5 A0A0P5VXP3 A0A0P5WXM8 A0A0P5JXK7 A0A0P5CWM3 A0A2H1VRA0 B4L403 A0A0P6CTY6 A0A0P5SV44 E9FXZ7 A0A0P5BKE4 E2BU28 A0A0P5HM40 A0A0P5N0G2 A0A0P5VKZ5 D6WT64
Ontologies
GO
PANTHER
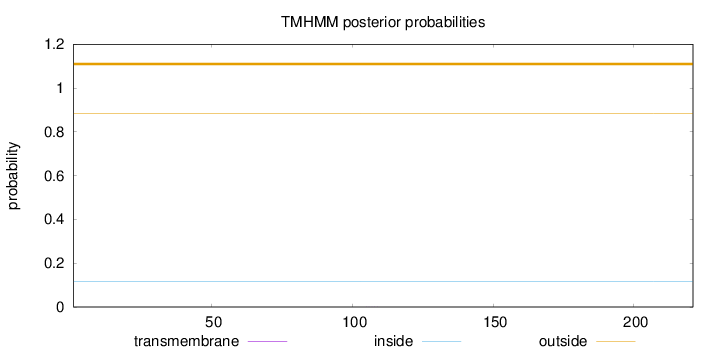
Topology
Subcellular location
Length:
221
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00364
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11539
outside
1 - 221
Population Genetic Test Statistics
Pi
42.903031
Theta
164.767095
Tajima's D
-1.651911
CLR
377.958174
CSRT
0.0443477826108695
Interpretation
Possibly Positive selection
