Pre Gene Modal
BGIBMGA009248
Annotation
PREDICTED:_T-complex_protein_11-like_protein_1_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.011 Golgi Reliability : 1.573
Sequence
CDS
ATGCACAGAGCGTACTTTGACATTTTGAGAGAACAGCTAAATTCCAATCCTCCTGAATACAAACAAGCCTTAATATTACTTGAAGACGTCAAACAGGGTCTCTTCTCAATATTATTACCAAGACATACTCGCATCCGTGAGATGATTGAGGAGGTCATAGATGCAGACTTCATAAAACAACAAGCGGAGACCAACAGTCTGGACTTTAAGAAATATGCCAACTTCGTTGTAGACCTCATGGCCAAATTAGCTGCCCCAGCAAGAGACGAAATGATACAAAACATTACTACACTCACCGATACTGTAGAGATCTTTCGAGCTATCCTAGAAGCACTTGAAGTACTGAAGCTAGATCTGGCCAATACATTGATCACTATGATCCGACCTCACGTTCAGAAAGAAAGTGTCCAGTACGAACGAGCTAAGTTTGATGAAATGCTTAAAGTTTCAGAGGACGGCCTTCAAAATACTAGGGAGTGGTTAAAACGACACATCGACAAAACTGATCTCAGTATACCGGTCACTGATCAGAACATAATCCGGAACGTCACCGCCCAAACCTTAGCCAAGGCATACCTAGAGCTGTTGGAGTGGGATTCGAGTAAAGGGTACCCGGAGACGGCGGCCATAGACGCGCCCCGGTTCTCTGAGCTGGGGGTGCAAGTGTACCGGATGTCGTGCGCGGCCGCGCTCCTACTGGCCAGCCCCGCGTGCTCCAGCGACGCCGACACCGCCGCGCGACACAAAGAAGCCCTCAAACAGAAGCTCTTCATCATCATCGACGAGGCCTCCAATGATACAGAACTAAGATCGGTGCTGCCATCTGTCGCCGAGGAGGTTGTATTAGCAACGGAACAACTGCTAGAAAAGTTGAACCAGCCCCCGCTGAACGCAGAGACGAAGGAACTCATACGAACTCAAATAATCTCCCTGGGTGATCCCGAGCATAGAGTTAGAGAGATTGTCCGTCAACGGGTTCTGGAGTTTCTGAAGGTCATACTGGTGTGCGGCGGAGGATCCCGTCAGATACCCGTCGGCCTGTCGGTGTTGTCGCGGGAACTCACCGACGTCTCCGGGACCCTGCTGCGGTACGTCATGCACAACAAAGCGGTCTTCACGCAGCACTACCTCGACATCGTCGCCGAGGAGTTGAGTAGGAACTGA
Protein
MHRAYFDILREQLNSNPPEYKQALILLEDVKQGLFSILLPRHTRIREMIEEVIDADFIKQQAETNSLDFKKYANFVVDLMAKLAAPARDEMIQNITTLTDTVEIFRAILEALEVLKLDLANTLITMIRPHVQKESVQYERAKFDEMLKVSEDGLQNTREWLKRHIDKTDLSIPVTDQNIIRNVTAQTLAKAYLELLEWDSSKGYPETAAIDAPRFSELGVQVYRMSCAAALLLASPACSSDADTAARHKEALKQKLFIIIDEASNDTELRSVLPSVAEEVVLATEQLLEKLNQPPLNAETKELIRTQIISLGDPEHRVREIVRQRVLEFLKVILVCGGGSRQIPVGLSVLSRELTDVSGTLLRYVMHNKAVFTQHYLDIVAEELSRN
Summary
Uniprot
H9JI98
A0A2H1V8B2
A0A194Q469
S4PS91
A0A212FHL5
A0A1W4XJT6
+ More
A0A2J7PCL5 A0A2J7PCK6 A0A2J7PCK9 D2A0W6 V5IA69 A0A1Y1KKC3 A0A0P4WUF6 E9FR38 A0A310S9A7 A0A154PJ74 A0A067QYV7 A0A087ZPX3 A0A2A3EJT9 A0A164ZIM7 A0A0P5GKY2 A0A0M8ZNK5 A0A0P5FUU7 A0A0P5XDI5 A0A0P6J4J3 A0A0L7RJP7 A0A0P5GIS1 A0A444SID1 A0A0P5HUU3 A0A1B6FRJ0 A0A1B6JZW6 E2C017 A0A3R7MS48 U4TY78 A0A443RJ00 T1IQ07 A0A1B6KLL7 A0A444SID2 A0A1E1XAG3 A0A151X269 A0A2S2QGP0 A0A026WE15 A0A0P5F710 A0A443STB3 A0A3L8E044 A0A195FJX4 A0A1Z5LF90 K7J2M7 E2AJX7 N6TT98 N6URB6 A0A232F764 F4W6K6 A0A195E4X5 A0A158P2B1 A0A293MVT3 A0A0P6DKQ6 R7VJ98 A0A0J7KNT5 B7Q963 A0A1S3IAZ9 A0A1S3IJZ7 A0A210Q234 A0A1B6DRJ1 A0A2S2PE60 A0A2R5LBU2 A0A224Z8Y9 A0A131XDZ5 E0VBB1 A0A224Z8A9 A0A2H8TPH6 A0A131XWZ1 L7M890 A0A0P5TIR4 T1KRM8 A0A0B7AL99 A0A0P5ZJU1 A0A0A1X7Y2 W8BJT7 A0A2T7P5S4 A0A0K8VU96 A0A0K8W529 A0A0L0BY74 A0A3M7PQ56 A0A034VF33 A0A146LJJ9 H0VVH3 A0A0K8TRG9 A0A0A9W663 A0A1B0BLH9 U6DDG7 S7P8T4 A0A1A9YQC6 G5AN91 D3TNT6 A0A1B0A1D8 A0A0D9QZ12 A0A2K5DQ70 A0A2K5H9L9
A0A2J7PCL5 A0A2J7PCK6 A0A2J7PCK9 D2A0W6 V5IA69 A0A1Y1KKC3 A0A0P4WUF6 E9FR38 A0A310S9A7 A0A154PJ74 A0A067QYV7 A0A087ZPX3 A0A2A3EJT9 A0A164ZIM7 A0A0P5GKY2 A0A0M8ZNK5 A0A0P5FUU7 A0A0P5XDI5 A0A0P6J4J3 A0A0L7RJP7 A0A0P5GIS1 A0A444SID1 A0A0P5HUU3 A0A1B6FRJ0 A0A1B6JZW6 E2C017 A0A3R7MS48 U4TY78 A0A443RJ00 T1IQ07 A0A1B6KLL7 A0A444SID2 A0A1E1XAG3 A0A151X269 A0A2S2QGP0 A0A026WE15 A0A0P5F710 A0A443STB3 A0A3L8E044 A0A195FJX4 A0A1Z5LF90 K7J2M7 E2AJX7 N6TT98 N6URB6 A0A232F764 F4W6K6 A0A195E4X5 A0A158P2B1 A0A293MVT3 A0A0P6DKQ6 R7VJ98 A0A0J7KNT5 B7Q963 A0A1S3IAZ9 A0A1S3IJZ7 A0A210Q234 A0A1B6DRJ1 A0A2S2PE60 A0A2R5LBU2 A0A224Z8Y9 A0A131XDZ5 E0VBB1 A0A224Z8A9 A0A2H8TPH6 A0A131XWZ1 L7M890 A0A0P5TIR4 T1KRM8 A0A0B7AL99 A0A0P5ZJU1 A0A0A1X7Y2 W8BJT7 A0A2T7P5S4 A0A0K8VU96 A0A0K8W529 A0A0L0BY74 A0A3M7PQ56 A0A034VF33 A0A146LJJ9 H0VVH3 A0A0K8TRG9 A0A0A9W663 A0A1B0BLH9 U6DDG7 S7P8T4 A0A1A9YQC6 G5AN91 D3TNT6 A0A1B0A1D8 A0A0D9QZ12 A0A2K5DQ70 A0A2K5H9L9
Pubmed
19121390
26354079
23622113
22118469
18362917
19820115
+ More
28004739 21292972 24845553 20798317 23537049 28503490 24508170 30249741 28528879 20075255 28648823 21719571 21347285 23254933 26383154 28812685 28797301 28049606 20566863 25576852 25830018 24495485 26108605 30375419 25348373 26823975 21993624 26369729 25401762 21993625 20353571
28004739 21292972 24845553 20798317 23537049 28503490 24508170 30249741 28528879 20075255 28648823 21719571 21347285 23254933 26383154 28812685 28797301 28049606 20566863 25576852 25830018 24495485 26108605 30375419 25348373 26823975 21993624 26369729 25401762 21993625 20353571
EMBL
BABH01022212
ODYU01000952
SOQ36484.1
KQ459585
KPI98205.1
GAIX01014058
+ More
JAA78502.1 AGBW02008491 OWR53233.1 NEVH01027059 PNF14069.1 PNF14068.1 PNF14070.1 KQ971338 EFA01622.1 GALX01001705 JAB66761.1 GEZM01083954 JAV60911.1 GDRN01058396 JAI65604.1 GL732523 EFX90259.1 KQ763209 OAD55150.1 KQ434910 KZC11348.1 KK852868 KDR14701.1 KZ288227 PBC31744.1 LRGB01000723 KZS16414.1 GDIQ01242885 JAK08840.1 KQ435966 KOX67940.1 GDIQ01251247 JAK00478.1 GDIP01075256 JAM28459.1 GDIQ01013347 JAN81390.1 KQ414579 KOC71095.1 GDIQ01244092 JAK07633.1 SAUD01035549 RXG53167.1 GDIQ01245447 JAK06278.1 GECZ01016937 JAS52832.1 GECU01002957 JAT04750.1 GL451712 EFN78713.1 QCYY01002665 ROT68448.1 KB631829 ERL86574.1 NCKU01000506 RWS15256.1 JH431273 GEBQ01027638 JAT12339.1 RXG53168.1 GFAC01002961 JAT96227.1 KQ982580 KYQ54507.1 GGMS01007726 MBY76929.1 KK107250 EZA54345.1 GDIQ01267020 JAJ84704.1 NCKV01000394 RWS30740.1 QOIP01000002 RLU25922.1 KQ981523 KYN40304.1 GFJQ02000904 JAW06066.1 GL440100 EFN66293.1 APGK01019793 KB740125 ENN81298.1 ENN81297.1 NNAY01000831 OXU26289.1 GL887707 EGI70341.1 KQ979657 KYN19952.1 ADTU01007051 GFWV01019333 MAA44061.1 GDIQ01090938 JAN03799.1 AMQN01000491 KB291799 ELU18704.1 LBMM01004880 KMQ92023.1 ABJB010817198 DS887628 EEC15385.1 NEDP02005224 OWF42739.1 GEDC01031353 GEDC01008994 GEDC01007784 JAS05945.1 JAS28304.1 JAS29514.1 GGMR01014859 MBY27478.1 GGLE01002799 MBY06925.1 GFPF01011918 MAA23064.1 GEFH01004283 JAP64298.1 DS235023 EEB10667.1 GFPF01011917 MAA23063.1 GFXV01004016 MBW15821.1 GEFM01004013 JAP71783.1 GACK01005705 JAA59329.1 GDIP01127249 JAL76465.1 CAEY01000414 HACG01034758 CEK81623.1 GDIP01042949 JAM60766.1 GBXI01006883 GBXI01003043 JAD07409.1 JAD11249.1 GAMC01016671 JAB89884.1 PZQS01000006 PVD28775.1 GDHF01024368 GDHF01009863 JAI27946.1 JAI42451.1 GDHF01025969 GDHF01006314 JAI26345.1 JAI46000.1 JRES01001250 KNC24234.1 REGN01009453 RNA01134.1 GAKP01018270 GAKP01018269 JAC40682.1 GDHC01010345 JAQ08284.1 AAKN02049891 GDAI01000641 JAI16962.1 GBHO01043249 JAG00355.1 JXJN01016357 HAAF01008837 CCP80661.1 KE162024 EPQ06643.1 JH166126 GEBF01004157 EHA98501.1 JAN99475.1 CCAG010001428 EZ423088 ADD19364.1 AQIB01084934
JAA78502.1 AGBW02008491 OWR53233.1 NEVH01027059 PNF14069.1 PNF14068.1 PNF14070.1 KQ971338 EFA01622.1 GALX01001705 JAB66761.1 GEZM01083954 JAV60911.1 GDRN01058396 JAI65604.1 GL732523 EFX90259.1 KQ763209 OAD55150.1 KQ434910 KZC11348.1 KK852868 KDR14701.1 KZ288227 PBC31744.1 LRGB01000723 KZS16414.1 GDIQ01242885 JAK08840.1 KQ435966 KOX67940.1 GDIQ01251247 JAK00478.1 GDIP01075256 JAM28459.1 GDIQ01013347 JAN81390.1 KQ414579 KOC71095.1 GDIQ01244092 JAK07633.1 SAUD01035549 RXG53167.1 GDIQ01245447 JAK06278.1 GECZ01016937 JAS52832.1 GECU01002957 JAT04750.1 GL451712 EFN78713.1 QCYY01002665 ROT68448.1 KB631829 ERL86574.1 NCKU01000506 RWS15256.1 JH431273 GEBQ01027638 JAT12339.1 RXG53168.1 GFAC01002961 JAT96227.1 KQ982580 KYQ54507.1 GGMS01007726 MBY76929.1 KK107250 EZA54345.1 GDIQ01267020 JAJ84704.1 NCKV01000394 RWS30740.1 QOIP01000002 RLU25922.1 KQ981523 KYN40304.1 GFJQ02000904 JAW06066.1 GL440100 EFN66293.1 APGK01019793 KB740125 ENN81298.1 ENN81297.1 NNAY01000831 OXU26289.1 GL887707 EGI70341.1 KQ979657 KYN19952.1 ADTU01007051 GFWV01019333 MAA44061.1 GDIQ01090938 JAN03799.1 AMQN01000491 KB291799 ELU18704.1 LBMM01004880 KMQ92023.1 ABJB010817198 DS887628 EEC15385.1 NEDP02005224 OWF42739.1 GEDC01031353 GEDC01008994 GEDC01007784 JAS05945.1 JAS28304.1 JAS29514.1 GGMR01014859 MBY27478.1 GGLE01002799 MBY06925.1 GFPF01011918 MAA23064.1 GEFH01004283 JAP64298.1 DS235023 EEB10667.1 GFPF01011917 MAA23063.1 GFXV01004016 MBW15821.1 GEFM01004013 JAP71783.1 GACK01005705 JAA59329.1 GDIP01127249 JAL76465.1 CAEY01000414 HACG01034758 CEK81623.1 GDIP01042949 JAM60766.1 GBXI01006883 GBXI01003043 JAD07409.1 JAD11249.1 GAMC01016671 JAB89884.1 PZQS01000006 PVD28775.1 GDHF01024368 GDHF01009863 JAI27946.1 JAI42451.1 GDHF01025969 GDHF01006314 JAI26345.1 JAI46000.1 JRES01001250 KNC24234.1 REGN01009453 RNA01134.1 GAKP01018270 GAKP01018269 JAC40682.1 GDHC01010345 JAQ08284.1 AAKN02049891 GDAI01000641 JAI16962.1 GBHO01043249 JAG00355.1 JXJN01016357 HAAF01008837 CCP80661.1 KE162024 EPQ06643.1 JH166126 GEBF01004157 EHA98501.1 JAN99475.1 CCAG010001428 EZ423088 ADD19364.1 AQIB01084934
Proteomes
UP000005204
UP000053268
UP000007151
UP000192223
UP000235965
UP000007266
+ More
UP000000305 UP000076502 UP000027135 UP000005203 UP000242457 UP000076858 UP000053105 UP000053825 UP000288706 UP000008237 UP000283509 UP000030742 UP000285301 UP000075809 UP000053097 UP000288716 UP000279307 UP000078541 UP000002358 UP000000311 UP000019118 UP000215335 UP000007755 UP000078492 UP000005205 UP000014760 UP000036403 UP000001555 UP000085678 UP000242188 UP000009046 UP000015104 UP000245119 UP000037069 UP000276133 UP000005447 UP000092460 UP000092443 UP000006813 UP000092444 UP000092445 UP000029965 UP000233020 UP000233080
UP000000305 UP000076502 UP000027135 UP000005203 UP000242457 UP000076858 UP000053105 UP000053825 UP000288706 UP000008237 UP000283509 UP000030742 UP000285301 UP000075809 UP000053097 UP000288716 UP000279307 UP000078541 UP000002358 UP000000311 UP000019118 UP000215335 UP000007755 UP000078492 UP000005205 UP000014760 UP000036403 UP000001555 UP000085678 UP000242188 UP000009046 UP000015104 UP000245119 UP000037069 UP000276133 UP000005447 UP000092460 UP000092443 UP000006813 UP000092444 UP000092445 UP000029965 UP000233020 UP000233080
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JI98
A0A2H1V8B2
A0A194Q469
S4PS91
A0A212FHL5
A0A1W4XJT6
+ More
A0A2J7PCL5 A0A2J7PCK6 A0A2J7PCK9 D2A0W6 V5IA69 A0A1Y1KKC3 A0A0P4WUF6 E9FR38 A0A310S9A7 A0A154PJ74 A0A067QYV7 A0A087ZPX3 A0A2A3EJT9 A0A164ZIM7 A0A0P5GKY2 A0A0M8ZNK5 A0A0P5FUU7 A0A0P5XDI5 A0A0P6J4J3 A0A0L7RJP7 A0A0P5GIS1 A0A444SID1 A0A0P5HUU3 A0A1B6FRJ0 A0A1B6JZW6 E2C017 A0A3R7MS48 U4TY78 A0A443RJ00 T1IQ07 A0A1B6KLL7 A0A444SID2 A0A1E1XAG3 A0A151X269 A0A2S2QGP0 A0A026WE15 A0A0P5F710 A0A443STB3 A0A3L8E044 A0A195FJX4 A0A1Z5LF90 K7J2M7 E2AJX7 N6TT98 N6URB6 A0A232F764 F4W6K6 A0A195E4X5 A0A158P2B1 A0A293MVT3 A0A0P6DKQ6 R7VJ98 A0A0J7KNT5 B7Q963 A0A1S3IAZ9 A0A1S3IJZ7 A0A210Q234 A0A1B6DRJ1 A0A2S2PE60 A0A2R5LBU2 A0A224Z8Y9 A0A131XDZ5 E0VBB1 A0A224Z8A9 A0A2H8TPH6 A0A131XWZ1 L7M890 A0A0P5TIR4 T1KRM8 A0A0B7AL99 A0A0P5ZJU1 A0A0A1X7Y2 W8BJT7 A0A2T7P5S4 A0A0K8VU96 A0A0K8W529 A0A0L0BY74 A0A3M7PQ56 A0A034VF33 A0A146LJJ9 H0VVH3 A0A0K8TRG9 A0A0A9W663 A0A1B0BLH9 U6DDG7 S7P8T4 A0A1A9YQC6 G5AN91 D3TNT6 A0A1B0A1D8 A0A0D9QZ12 A0A2K5DQ70 A0A2K5H9L9
A0A2J7PCL5 A0A2J7PCK6 A0A2J7PCK9 D2A0W6 V5IA69 A0A1Y1KKC3 A0A0P4WUF6 E9FR38 A0A310S9A7 A0A154PJ74 A0A067QYV7 A0A087ZPX3 A0A2A3EJT9 A0A164ZIM7 A0A0P5GKY2 A0A0M8ZNK5 A0A0P5FUU7 A0A0P5XDI5 A0A0P6J4J3 A0A0L7RJP7 A0A0P5GIS1 A0A444SID1 A0A0P5HUU3 A0A1B6FRJ0 A0A1B6JZW6 E2C017 A0A3R7MS48 U4TY78 A0A443RJ00 T1IQ07 A0A1B6KLL7 A0A444SID2 A0A1E1XAG3 A0A151X269 A0A2S2QGP0 A0A026WE15 A0A0P5F710 A0A443STB3 A0A3L8E044 A0A195FJX4 A0A1Z5LF90 K7J2M7 E2AJX7 N6TT98 N6URB6 A0A232F764 F4W6K6 A0A195E4X5 A0A158P2B1 A0A293MVT3 A0A0P6DKQ6 R7VJ98 A0A0J7KNT5 B7Q963 A0A1S3IAZ9 A0A1S3IJZ7 A0A210Q234 A0A1B6DRJ1 A0A2S2PE60 A0A2R5LBU2 A0A224Z8Y9 A0A131XDZ5 E0VBB1 A0A224Z8A9 A0A2H8TPH6 A0A131XWZ1 L7M890 A0A0P5TIR4 T1KRM8 A0A0B7AL99 A0A0P5ZJU1 A0A0A1X7Y2 W8BJT7 A0A2T7P5S4 A0A0K8VU96 A0A0K8W529 A0A0L0BY74 A0A3M7PQ56 A0A034VF33 A0A146LJJ9 H0VVH3 A0A0K8TRG9 A0A0A9W663 A0A1B0BLH9 U6DDG7 S7P8T4 A0A1A9YQC6 G5AN91 D3TNT6 A0A1B0A1D8 A0A0D9QZ12 A0A2K5DQ70 A0A2K5H9L9
Ontologies
PANTHER
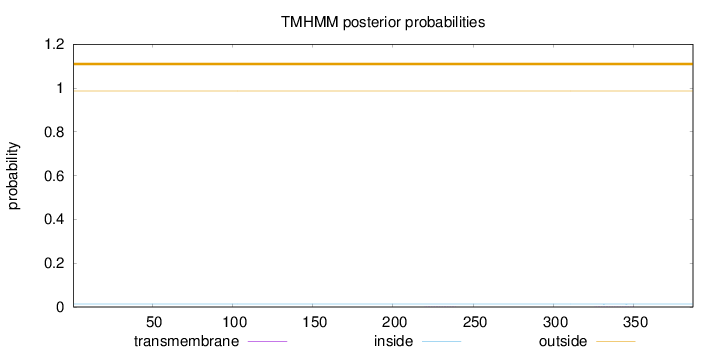
Topology
Length:
387
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01334
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.01354
outside
1 - 387
Population Genetic Test Statistics
Pi
182.954066
Theta
146.124793
Tajima's D
-0.690957
CLR
176.082498
CSRT
0.2000399980001
Interpretation
Uncertain
