Gene
KWMTBOMO08310 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009249
Annotation
nucleosome_assembly_protein_isoform_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.786
Sequence
CDS
ATGGGTACCGTCGAACGTTCTGGTGACGACCACACATCCGAAATGGAGTCTGCAGAAGAAGAAGAAGTCGTCGGAAGTGGTGAACTAGCGCAACACCTGTTGAAAAGTGGCGTGACCCGTAATGAAATGATAGCGGCCATCACAAATCGCCTTCATGCAGAAGCGATGGCATCCCTACCCCCGAATGTTCGTCGGCGAATCCGCGCCTTGAGAACTCTTCAGAAGGAGTTTGTCGACATTGAGGCCAAGTTTTACAGTGAAGTACATGCACTCGAATGCAAATATGAAAAACTTTACAAGCCTCTTTATGAAAAGCGAGCTCTTATTGTGAATGGCACATATGAACCTAATGATGATGAATGTCTCAACCCATGGCGTGATGACACTGAAGAAGAAGAGTTAGCTCGGGCGGTACAAAATGCTGCCATCACTGAGGGTGAGGAAAAGAAGGATGACAAGGCTATCGAGCCTCCAATGGATCCCAATGTAAAGGGTATCCCAGACTTTTGGTACAACATATTCAGGAATGTCTCAATGCTCAGCGAAATGATGCAAGAACATGATGAACCAATTCTAAAATGCTTGCAAGATATTAAAGTGCAAATGCATGAAGACCCCATAAGCTTCACTCTGGAATTCTACTTTGCTCCAAATGAATACTTCACAAATACAGTACTTACTAAAGAGTACTTAATGAAGTGCAAGCCCGATGAAGAGAGTCCATTAGAATTTGAAGGTCCTGAAATTTATTCTTGCAAGGGATGTGAAATAAACTGGAATGCTGGTAAAAACGTAACTGTCAAGACCATAAAGAAGAAACAGAAGCACAAGTCCCGTGGGTCAGTTCGCACTGTCACTAAGTCCGTCCAAGCTGACTCCTTCTTCAATTTCTTTAACCCACCAACTCTGCCGGAAGATCCTAACTCTACAGTCGCCTCTGATGTACAGGCGTTGTTGACTGCGGACTTTGAGATCGGTCACTACATCCGCGAGCGCGTCGTGTCTCGTGCCGTGCTGCTCTACACCGGCGAGGGACTCGAGGATGATGACGATGACGATTATGAAGAAGAGGAGGAAGAGGAATGCTCGATGGAGGAGAGCGAAGACGATGAGCCGGCGCCTCGGAGACGAACCAAGAAACCCGGCAAGGGACCTGCCGAGAACCCCGCTGAGTGCAAACAGCAGTAG
Protein
MGTVERSGDDHTSEMESAEEEEVVGSGELAQHLLKSGVTRNEMIAAITNRLHAEAMASLPPNVRRRIRALRTLQKEFVDIEAKFYSEVHALECKYEKLYKPLYEKRALIVNGTYEPNDDECLNPWRDDTEEEELARAVQNAAITEGEEKKDDKAIEPPMDPNVKGIPDFWYNIFRNVSMLSEMMQEHDEPILKCLQDIKVQMHEDPISFTLEFYFAPNEYFTNTVLTKEYLMKCKPDEESPLEFEGPEIYSCKGCEINWNAGKNVTVKTIKKKQKHKSRGSVRTVTKSVQADSFFNFFNPPTLPEDPNSTVASDVQALLTADFEIGHYIRERVVSRAVLLYTGEGLEDDDDDDYEEEEEEECSMEESEDDEPAPRRRTKKPGKGPAENPAECKQQ
Summary
Similarity
Belongs to the nucleosome assembly protein (NAP) family.
Uniprot
Q1HQB9
Q2F5X9
H9JI99
A0A2H1V7V8
A0A3S2TRD3
A0A1E1W5B4
+ More
S4PGN1 A0A212FHM0 B2DBN5 A0A194PYU4 A0A194R6M7 A0A0L7LP29 A0A067RSE1 A0A1B6LNY8 A0A1B6MB76 A0A310SN27 A0A423T1L6 A0A2J7PYL5 A0A1Y1LRB3 A0A0C9Q9S3 A0A411G838 A0A158NEK9 A0A0P4WGJ0 A0A2A3EKN8 V9IIC1 A0A154PRH6 A0A026X0C5 A0A195BWN0 A0A195EN10 A0A1B6DHR9 A0A195BXX5 F4WRV1 A0A3L8DGQ4 A0A151XIS7 A0A195FLB5 A0A1W4XL05 A0A0N0U6S9 E2AFN4 E9ID94 A0A232EU00 A0A1D1Y7D0 A0A0J7LA52 E2B3E6 A0A139WJ25 A0A0P5ENK8 A0A0L7R6B2 N6SUP9 A0A088A460 A0A0P5XA12 A0A0P5G3I4 J3JUD3 E9FRI1 A0A0P5EX38 A0A0P5NP99 A0A0P5JS38 A0A0P4YE19 A0A0P5IRT0 T1JKL6 A0A0P6ACX3 A0A0N8C5X1 A0A0P6CM42 A0A2P2HXV3 A0A0P5MV73 A0A0P5R3N5 A0A0P6I1S4 A0A0P5H472 E0V9A3 A0A1B6EW74 A0A0P5HIG5 A0A1B6L4H3 A0A0P6GH46 A0A0P5IF91 A0A2H8THB4 A0A3S1A0E0 A0A0V0G4G5 A0A0P6FGB9 A0A224XIG6 A0A2S2NEB5 J9JVG1 A0A069DSH5 A0A0P4ZQJ7 A0A444TUC1 T1IA74 K1QKW8 A0A0P5RLE1 A0A0P5QQ17 A0A210QUG5 W5LLG5 W5LLE7 A0A0P5R7B9 A0A444SZ81 A0A0P5FPM2 H2RPU0 A0A3P9JQU9 A0A3P9IIV3 A0A1A8M8U4 A0A1A8PQ69 A0A1A8HWZ6 A0A1A8D251
S4PGN1 A0A212FHM0 B2DBN5 A0A194PYU4 A0A194R6M7 A0A0L7LP29 A0A067RSE1 A0A1B6LNY8 A0A1B6MB76 A0A310SN27 A0A423T1L6 A0A2J7PYL5 A0A1Y1LRB3 A0A0C9Q9S3 A0A411G838 A0A158NEK9 A0A0P4WGJ0 A0A2A3EKN8 V9IIC1 A0A154PRH6 A0A026X0C5 A0A195BWN0 A0A195EN10 A0A1B6DHR9 A0A195BXX5 F4WRV1 A0A3L8DGQ4 A0A151XIS7 A0A195FLB5 A0A1W4XL05 A0A0N0U6S9 E2AFN4 E9ID94 A0A232EU00 A0A1D1Y7D0 A0A0J7LA52 E2B3E6 A0A139WJ25 A0A0P5ENK8 A0A0L7R6B2 N6SUP9 A0A088A460 A0A0P5XA12 A0A0P5G3I4 J3JUD3 E9FRI1 A0A0P5EX38 A0A0P5NP99 A0A0P5JS38 A0A0P4YE19 A0A0P5IRT0 T1JKL6 A0A0P6ACX3 A0A0N8C5X1 A0A0P6CM42 A0A2P2HXV3 A0A0P5MV73 A0A0P5R3N5 A0A0P6I1S4 A0A0P5H472 E0V9A3 A0A1B6EW74 A0A0P5HIG5 A0A1B6L4H3 A0A0P6GH46 A0A0P5IF91 A0A2H8THB4 A0A3S1A0E0 A0A0V0G4G5 A0A0P6FGB9 A0A224XIG6 A0A2S2NEB5 J9JVG1 A0A069DSH5 A0A0P4ZQJ7 A0A444TUC1 T1IA74 K1QKW8 A0A0P5RLE1 A0A0P5QQ17 A0A210QUG5 W5LLG5 W5LLE7 A0A0P5R7B9 A0A444SZ81 A0A0P5FPM2 H2RPU0 A0A3P9JQU9 A0A3P9IIV3 A0A1A8M8U4 A0A1A8PQ69 A0A1A8HWZ6 A0A1A8D251
Pubmed
EMBL
DQ443133
ABF51222.1
DQ311294
ABD36238.1
BABH01022211
ODYU01000952
+ More
SOQ36482.1 RSAL01000016 RVE53047.1 GDQN01008881 GDQN01004190 JAT82173.1 JAT86864.1 GAIX01003587 JAA88973.1 AGBW02008491 OWR53231.1 AB264720 BAG30794.1 KQ459585 KPI98203.1 KQ460627 KPJ13468.1 JTDY01000452 KOB77114.1 KK852446 KDR23730.1 GEBQ01014564 JAT25413.1 GEBQ01006791 JAT33186.1 KQ762962 OAD55188.1 QCYY01002425 ROT70439.1 NEVH01020348 PNF21402.1 GEZM01052548 JAV74445.1 GBYB01011123 JAG80890.1 MH366291 QBB02100.1 ADTU01013487 GDRN01015154 JAI67851.1 KZ288219 PBC32287.1 JR047363 AEY60392.1 KQ435066 KZC14337.1 KK107064 EZA60849.1 KQ976401 KYM92378.1 KQ978625 KYN29588.1 GEDC01012065 JAS25233.1 KQ978501 KYM93447.1 GL888292 EGI63108.1 QOIP01000008 RLU19501.1 KQ982080 KYQ60316.1 KQ981490 KYN41203.1 KQ435728 KOX77758.1 GL439118 EFN67785.1 GL762454 EFZ21423.1 NNAY01002197 OXU21831.1 GDJX01017381 JAT50555.1 LBMM01000137 KMR04792.1 GL445323 EFN89824.1 KQ971338 KYB27922.1 GDIQ01267222 JAJ84502.1 KQ414648 KOC66316.1 APGK01055882 APGK01055883 KB741270 ENN71414.1 GDIP01087305 JAM16410.1 GDIQ01247005 JAK04720.1 BT126846 AEE61808.1 GL732523 EFX90170.1 GDIP01154501 LRGB01013583 JAJ68901.1 KZR99454.1 GDIQ01146334 JAL05392.1 GDIQ01195853 JAK55872.1 GDIP01231552 JAI91849.1 GDIQ01217752 JAK33973.1 JH432078 GDIP01036113 JAM67602.1 GDIQ01111691 JAL40035.1 GDIP01000131 JAN03585.1 IACF01000858 LAB66601.1 GDIQ01162129 JAK89596.1 GDIQ01111692 JAL40034.1 GDIQ01010758 JAN83979.1 GDIQ01232952 JAK18773.1 DS234991 EEB09959.1 GECZ01027533 JAS42236.1 GDIQ01232953 JAK18772.1 GEBQ01021446 JAT18531.1 GDIQ01045722 JAN49015.1 GDIQ01213257 JAK38468.1 GFXV01000853 MBW12658.1 RQTK01000109 RUS87503.1 GECL01003222 JAP02902.1 GDIQ01049021 JAN45716.1 GFTR01005582 JAW10844.1 GGMR01002829 MBY15448.1 ABLF02039073 GBGD01001851 JAC87038.1 GDIP01211096 JAJ12306.1 SAUD01000314 RXG73541.1 ACPB03005987 JH816918 EKC37417.1 GDIQ01099085 JAL52641.1 GDIQ01111693 JAL40033.1 NEDP02001812 OWF52380.1 GDIQ01111694 JAL40032.1 SAUD01018030 RXG58663.1 GDIQ01251749 JAJ99975.1 HAEF01012083 SBR53242.1 HAEH01007926 SBR83147.1 HAED01003070 HAEE01004726 SBQ88959.1 HADZ01021264 HAEA01000461 SBP85205.1
SOQ36482.1 RSAL01000016 RVE53047.1 GDQN01008881 GDQN01004190 JAT82173.1 JAT86864.1 GAIX01003587 JAA88973.1 AGBW02008491 OWR53231.1 AB264720 BAG30794.1 KQ459585 KPI98203.1 KQ460627 KPJ13468.1 JTDY01000452 KOB77114.1 KK852446 KDR23730.1 GEBQ01014564 JAT25413.1 GEBQ01006791 JAT33186.1 KQ762962 OAD55188.1 QCYY01002425 ROT70439.1 NEVH01020348 PNF21402.1 GEZM01052548 JAV74445.1 GBYB01011123 JAG80890.1 MH366291 QBB02100.1 ADTU01013487 GDRN01015154 JAI67851.1 KZ288219 PBC32287.1 JR047363 AEY60392.1 KQ435066 KZC14337.1 KK107064 EZA60849.1 KQ976401 KYM92378.1 KQ978625 KYN29588.1 GEDC01012065 JAS25233.1 KQ978501 KYM93447.1 GL888292 EGI63108.1 QOIP01000008 RLU19501.1 KQ982080 KYQ60316.1 KQ981490 KYN41203.1 KQ435728 KOX77758.1 GL439118 EFN67785.1 GL762454 EFZ21423.1 NNAY01002197 OXU21831.1 GDJX01017381 JAT50555.1 LBMM01000137 KMR04792.1 GL445323 EFN89824.1 KQ971338 KYB27922.1 GDIQ01267222 JAJ84502.1 KQ414648 KOC66316.1 APGK01055882 APGK01055883 KB741270 ENN71414.1 GDIP01087305 JAM16410.1 GDIQ01247005 JAK04720.1 BT126846 AEE61808.1 GL732523 EFX90170.1 GDIP01154501 LRGB01013583 JAJ68901.1 KZR99454.1 GDIQ01146334 JAL05392.1 GDIQ01195853 JAK55872.1 GDIP01231552 JAI91849.1 GDIQ01217752 JAK33973.1 JH432078 GDIP01036113 JAM67602.1 GDIQ01111691 JAL40035.1 GDIP01000131 JAN03585.1 IACF01000858 LAB66601.1 GDIQ01162129 JAK89596.1 GDIQ01111692 JAL40034.1 GDIQ01010758 JAN83979.1 GDIQ01232952 JAK18773.1 DS234991 EEB09959.1 GECZ01027533 JAS42236.1 GDIQ01232953 JAK18772.1 GEBQ01021446 JAT18531.1 GDIQ01045722 JAN49015.1 GDIQ01213257 JAK38468.1 GFXV01000853 MBW12658.1 RQTK01000109 RUS87503.1 GECL01003222 JAP02902.1 GDIQ01049021 JAN45716.1 GFTR01005582 JAW10844.1 GGMR01002829 MBY15448.1 ABLF02039073 GBGD01001851 JAC87038.1 GDIP01211096 JAJ12306.1 SAUD01000314 RXG73541.1 ACPB03005987 JH816918 EKC37417.1 GDIQ01099085 JAL52641.1 GDIQ01111693 JAL40033.1 NEDP02001812 OWF52380.1 GDIQ01111694 JAL40032.1 SAUD01018030 RXG58663.1 GDIQ01251749 JAJ99975.1 HAEF01012083 SBR53242.1 HAEH01007926 SBR83147.1 HAED01003070 HAEE01004726 SBQ88959.1 HADZ01021264 HAEA01000461 SBP85205.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000027135 UP000283509 UP000235965 UP000005205 UP000242457 UP000076502 UP000053097 UP000078540 UP000078492 UP000078542 UP000007755 UP000279307 UP000075809 UP000078541 UP000192223 UP000053105 UP000000311 UP000215335 UP000036403 UP000008237 UP000007266 UP000053825 UP000019118 UP000005203 UP000000305 UP000076858 UP000009046 UP000271974 UP000007819 UP000288706 UP000015103 UP000005408 UP000242188 UP000018467 UP000005226 UP000265200
UP000027135 UP000283509 UP000235965 UP000005205 UP000242457 UP000076502 UP000053097 UP000078540 UP000078492 UP000078542 UP000007755 UP000279307 UP000075809 UP000078541 UP000192223 UP000053105 UP000000311 UP000215335 UP000036403 UP000008237 UP000007266 UP000053825 UP000019118 UP000005203 UP000000305 UP000076858 UP000009046 UP000271974 UP000007819 UP000288706 UP000015103 UP000005408 UP000242188 UP000018467 UP000005226 UP000265200
Pfam
PF00956 NAP
SUPFAM
SSF143113
SSF143113
ProteinModelPortal
Q1HQB9
Q2F5X9
H9JI99
A0A2H1V7V8
A0A3S2TRD3
A0A1E1W5B4
+ More
S4PGN1 A0A212FHM0 B2DBN5 A0A194PYU4 A0A194R6M7 A0A0L7LP29 A0A067RSE1 A0A1B6LNY8 A0A1B6MB76 A0A310SN27 A0A423T1L6 A0A2J7PYL5 A0A1Y1LRB3 A0A0C9Q9S3 A0A411G838 A0A158NEK9 A0A0P4WGJ0 A0A2A3EKN8 V9IIC1 A0A154PRH6 A0A026X0C5 A0A195BWN0 A0A195EN10 A0A1B6DHR9 A0A195BXX5 F4WRV1 A0A3L8DGQ4 A0A151XIS7 A0A195FLB5 A0A1W4XL05 A0A0N0U6S9 E2AFN4 E9ID94 A0A232EU00 A0A1D1Y7D0 A0A0J7LA52 E2B3E6 A0A139WJ25 A0A0P5ENK8 A0A0L7R6B2 N6SUP9 A0A088A460 A0A0P5XA12 A0A0P5G3I4 J3JUD3 E9FRI1 A0A0P5EX38 A0A0P5NP99 A0A0P5JS38 A0A0P4YE19 A0A0P5IRT0 T1JKL6 A0A0P6ACX3 A0A0N8C5X1 A0A0P6CM42 A0A2P2HXV3 A0A0P5MV73 A0A0P5R3N5 A0A0P6I1S4 A0A0P5H472 E0V9A3 A0A1B6EW74 A0A0P5HIG5 A0A1B6L4H3 A0A0P6GH46 A0A0P5IF91 A0A2H8THB4 A0A3S1A0E0 A0A0V0G4G5 A0A0P6FGB9 A0A224XIG6 A0A2S2NEB5 J9JVG1 A0A069DSH5 A0A0P4ZQJ7 A0A444TUC1 T1IA74 K1QKW8 A0A0P5RLE1 A0A0P5QQ17 A0A210QUG5 W5LLG5 W5LLE7 A0A0P5R7B9 A0A444SZ81 A0A0P5FPM2 H2RPU0 A0A3P9JQU9 A0A3P9IIV3 A0A1A8M8U4 A0A1A8PQ69 A0A1A8HWZ6 A0A1A8D251
S4PGN1 A0A212FHM0 B2DBN5 A0A194PYU4 A0A194R6M7 A0A0L7LP29 A0A067RSE1 A0A1B6LNY8 A0A1B6MB76 A0A310SN27 A0A423T1L6 A0A2J7PYL5 A0A1Y1LRB3 A0A0C9Q9S3 A0A411G838 A0A158NEK9 A0A0P4WGJ0 A0A2A3EKN8 V9IIC1 A0A154PRH6 A0A026X0C5 A0A195BWN0 A0A195EN10 A0A1B6DHR9 A0A195BXX5 F4WRV1 A0A3L8DGQ4 A0A151XIS7 A0A195FLB5 A0A1W4XL05 A0A0N0U6S9 E2AFN4 E9ID94 A0A232EU00 A0A1D1Y7D0 A0A0J7LA52 E2B3E6 A0A139WJ25 A0A0P5ENK8 A0A0L7R6B2 N6SUP9 A0A088A460 A0A0P5XA12 A0A0P5G3I4 J3JUD3 E9FRI1 A0A0P5EX38 A0A0P5NP99 A0A0P5JS38 A0A0P4YE19 A0A0P5IRT0 T1JKL6 A0A0P6ACX3 A0A0N8C5X1 A0A0P6CM42 A0A2P2HXV3 A0A0P5MV73 A0A0P5R3N5 A0A0P6I1S4 A0A0P5H472 E0V9A3 A0A1B6EW74 A0A0P5HIG5 A0A1B6L4H3 A0A0P6GH46 A0A0P5IF91 A0A2H8THB4 A0A3S1A0E0 A0A0V0G4G5 A0A0P6FGB9 A0A224XIG6 A0A2S2NEB5 J9JVG1 A0A069DSH5 A0A0P4ZQJ7 A0A444TUC1 T1IA74 K1QKW8 A0A0P5RLE1 A0A0P5QQ17 A0A210QUG5 W5LLG5 W5LLE7 A0A0P5R7B9 A0A444SZ81 A0A0P5FPM2 H2RPU0 A0A3P9JQU9 A0A3P9IIV3 A0A1A8M8U4 A0A1A8PQ69 A0A1A8HWZ6 A0A1A8D251
PDB
6N2G
E-value=1.06684e-42,
Score=436
Ontologies
PANTHER
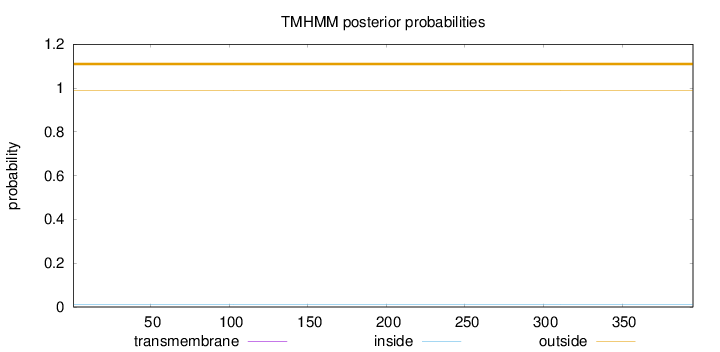
Topology
Length:
395
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00038
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.01017
outside
1 - 395
Population Genetic Test Statistics
Pi
169.201609
Theta
163.430671
Tajima's D
-0.382169
CLR
0.464937
CSRT
0.267986600669966
Interpretation
Uncertain
