Gene
KWMTBOMO08302
Pre Gene Modal
BGIBMGA009253
Annotation
PREDICTED:_protein_goliath_isoform_X1_[Papilio_xuthus]
Full name
Protein goliath
Alternative Name
Protein g1
Location in the cell
PlasmaMembrane Reliability : 2.255
Sequence
CDS
ATGCGTGCGGAAGAATTGTGGTCTGGTTTAGCGGCTCTGGGATGGATGGTTCTGGTGGTGAATTGCCATACGCCCGGGAGCGAACTGGAGACAGTACGTTACGGCCCGACAGATGAGCGCTTGTCCAAGACGATAGCGAATATCAACATAACGTACACTGACGAGCACGGAGTTGTGTATTCGGAAATATCGGAGTCCGGAAAATATGGCGAAGGATTCGTTGGGTCATCGAGCGGGGTCGGCGTGCACATCAGAGCCAAGGGCCCGGGCGGGGAAAGGGACCACACCGGCTGCTCGTGGCCCCTGCTGTCAACCACCGCGCCCAATGAACCTTTACCTCAAGAGCCCTGGATTGCTCTCATAAGACGAGGCAGCTGCAATTTTGAACTAAAGGTGCAGAACGCGTGGCGCGCCAACGCTACCGCAGTCCTTATATACAACGACCGCGATGCTCCAGATCTCGACAAAATGAAGCTGTCCACGGATAACGGACGTAATATAAGTGCCGTGTTCACGTACAAGTGGAAGGGAGAAGAGATAGCGCGTCTCATTGACAACGGCACCAGAGTCACCATCGCTATAATCAAAGGGAGGTCCTTCGCAATCATACCCAAAGACTTCAACAAGACATCAGTACTGTTCGTGTCTATATCTTTTATCGTGCTCATGGTTATATCATTGGCGTGGCTCGTCTTCTACTACATACAACGTTTCCGATATATACACGCCAAAGACAGGCTCTCTAAACGTCTGTGCTGCGCTGCGAAGAAAGCTTTATCTAAAATTCCCGTTAAGAGCTTAAAGACTGAAGACAGAGAAGTGCAAGGAGACGGCGAATGCTGTGCTATATGTATAGAGCCTTACAAAGTATCAGAAACATTGAGGTCTCTGCCGTGCAGGCACGATTTCCATAAGAACTGCATAGACCCGTGGCTGTTGGAGCATCGGACGTGTCCCATGTGCAAGATGGACATACTAAAGTACTACGGGTTTGTGTTTACTGGGAGTCAGGAGAGTATTCTGCAGTTAGAAGTGGAGGAAACGAGAGGCCACGCATTGAGTCCCTCTCGTAATAGACATCCATTAACTTTGTTAACGAGCGACAACTCTTACGACGAGGCGATCAGCGAACGAAGCAGTCGCGCTTCCTCGCCGGATCAACTGGTTCCGCGCTGTCCCCACGAGACCAGATCATGCACATCAACTCCATCCGGATCACCGCGACCAAATACCTCGCGACAACACGACGTCGCCTCCACCAGCACCTCCAACGCTCAGGAGTGA
Protein
MRAEELWSGLAALGWMVLVVNCHTPGSELETVRYGPTDERLSKTIANINITYTDEHGVVYSEISESGKYGEGFVGSSSGVGVHIRAKGPGGERDHTGCSWPLLSTTAPNEPLPQEPWIALIRRGSCNFELKVQNAWRANATAVLIYNDRDAPDLDKMKLSTDNGRNISAVFTYKWKGEEIARLIDNGTRVTIAIIKGRSFAIIPKDFNKTSVLFVSISFIVLMVISLAWLVFYYIQRFRYIHAKDRLSKRLCCAAKKALSKIPVKSLKTEDREVQGDGECCAICIEPYKVSETLRSLPCRHDFHKNCIDPWLLEHRTCPMCKMDILKYYGFVFTGSQESILQLEVEETRGHALSPSRNRHPLTLLTSDNSYDEAISERSSRASSPDQLVPRCPHETRSCTSTPSGSPRPNTSRQHDVASTSTSNAQE
Summary
Description
Regulation of gene expression during embryonic mesoderm formation. Putative role as transcription factor.
Keywords
Alternative splicing
Complete proteome
Developmental protein
DNA-binding
Membrane
Metal-binding
Reference proteome
Signal
Transcription
Transcription regulation
Transmembrane
Transmembrane helix
Zinc
Zinc-finger
Feature
chain Protein goliath
splice variant In isoform C.
splice variant In isoform C.
Uniprot
A0A3S2M767
A0A212EH97
A0A194RC27
A0A194PXX5
D2A1E8
A0A1B6DHR0
+ More
A0A1Y1M0X6 J9K827 A0A2S2QHA3 A0A1B6EDU2 A0A1B6E4H6 A0A1B6DIP4 A0A1B6F3T8 A0A1S4FVT7 A0A182V8R7 A0A453Z0L3 A0A1B6IGP3 A0A0P5Z6D6 A0A0P5YP19 A0A182F458 A0A1B6JTW9 E9HC82 A0A0P6GWM5 A0A1B6FJY8 A0A0P5BXF3 A0A2M3ZE25 A0A2M4BJT6 Q5TUG4 A0A023ETC3 A0A2M4CGZ8 A0A2M4CGF9 A0A2M4CGG0 A0A2M4AB69 A0A2M4ABB7 T1J3Y4 Q16MD5 A0A0P5EA85 A0A1Y9H9H6 A0A453Z0X3 A0A182HX16 A0A182PSD2 T1PJK6 A0A1I8MG78 A0A226E6X8 A0A182KGY3 A0A1Z5LFW6 A0A0J9RM83 A0A0J9RKN1 Q06003 A0A1Z5KU94 A0A1I8NSU6 A0A182R910 B3NRA2 A0A0A1XJ46 B4PBZ6 A0A293MZG4 A0A182W877 Q06003-4 B4KSS0 B4IHD0 A0A182YE34 A0A1W4V5W9 A0A3B0JHF5 B4LNU5 Q28Z87 T1PB96 A0A034VB16 V5H2L8 A0A131Y031 A0A0K8V8G7 A0A3Q0JKC6 A0A3B0IY81 B3MCD9 A0A1B0BGP2 Q06003-3 A0A0B4K8B7 A0A1A9WV64 B4GI37 B4MPL8 A0A0L0CC15 B4JVJ7 R7TQG9
A0A1Y1M0X6 J9K827 A0A2S2QHA3 A0A1B6EDU2 A0A1B6E4H6 A0A1B6DIP4 A0A1B6F3T8 A0A1S4FVT7 A0A182V8R7 A0A453Z0L3 A0A1B6IGP3 A0A0P5Z6D6 A0A0P5YP19 A0A182F458 A0A1B6JTW9 E9HC82 A0A0P6GWM5 A0A1B6FJY8 A0A0P5BXF3 A0A2M3ZE25 A0A2M4BJT6 Q5TUG4 A0A023ETC3 A0A2M4CGZ8 A0A2M4CGF9 A0A2M4CGG0 A0A2M4AB69 A0A2M4ABB7 T1J3Y4 Q16MD5 A0A0P5EA85 A0A1Y9H9H6 A0A453Z0X3 A0A182HX16 A0A182PSD2 T1PJK6 A0A1I8MG78 A0A226E6X8 A0A182KGY3 A0A1Z5LFW6 A0A0J9RM83 A0A0J9RKN1 Q06003 A0A1Z5KU94 A0A1I8NSU6 A0A182R910 B3NRA2 A0A0A1XJ46 B4PBZ6 A0A293MZG4 A0A182W877 Q06003-4 B4KSS0 B4IHD0 A0A182YE34 A0A1W4V5W9 A0A3B0JHF5 B4LNU5 Q28Z87 T1PB96 A0A034VB16 V5H2L8 A0A131Y031 A0A0K8V8G7 A0A3Q0JKC6 A0A3B0IY81 B3MCD9 A0A1B0BGP2 Q06003-3 A0A0B4K8B7 A0A1A9WV64 B4GI37 B4MPL8 A0A0L0CC15 B4JVJ7 R7TQG9
Pubmed
22118469
26354079
18362917
19820115
28004739
17510324
+ More
12364791 21292972 24945155 25315136 28528879 22936249 10731132 12537572 12537569 8462875 17994087 25830018 17550304 18057021 25244985 15632085 25348373 25765539 29652888 12537568 12537573 12537574 16110336 17569856 17569867 26108605 23254933
12364791 21292972 24945155 25315136 28528879 22936249 10731132 12537572 12537569 8462875 17994087 25830018 17550304 18057021 25244985 15632085 25348373 25765539 29652888 12537568 12537573 12537574 16110336 17569856 17569867 26108605 23254933
EMBL
RSAL01000016
RVE53042.1
AGBW02014927
OWR40852.1
KQ460627
KPJ13461.1
+ More
KQ459585 KPI98196.1 KQ971338 EFA01541.2 GEDC01012082 JAS25216.1 GEZM01045043 JAV77955.1 ABLF02018593 ABLF02018595 ABLF02019204 ABLF02019206 ABLF02019207 GGMS01007349 MBY76552.1 GEDC01001238 JAS36060.1 GEDC01031480 GEDC01004466 JAS05818.1 JAS32832.1 GEDC01011793 JAS25505.1 GECZ01024932 JAS44837.1 AAAB01008849 GECU01025788 GECU01021631 JAS81918.1 JAS86075.1 GDIP01048315 LRGB01001569 JAM55400.1 KZS11735.1 GDIP01055915 JAM47800.1 GECU01005087 JAT02620.1 GL732619 EFX70577.1 GDIQ01028417 JAN66320.1 GECZ01019278 JAS50491.1 GDIP01182642 JAJ40760.1 GGFM01006004 MBW26755.1 GGFJ01004195 MBW53336.1 EAL41047.3 GAPW01001045 JAC12553.1 GGFL01000253 MBW64431.1 GGFL01000254 MBW64432.1 GGFL01000252 MBW64430.1 GGFK01004713 MBW38034.1 GGFK01004729 MBW38050.1 JH431832 CH477865 EAT35498.1 GDIP01145996 JAJ77406.1 AXCN02002284 APCN01005116 KA648118 AFP62747.1 LNIX01000006 OXA52717.1 GFJQ02000677 JAW06293.1 CM002911 KMY96524.1 KMY96521.1 KMY96522.1 KMY96523.1 AE013599 AY069169 M97204 GFJQ02008307 JAV98662.1 CH954179 EDV57121.1 GBXI01002933 JAD11359.1 CM000158 EDW92650.1 KRK00624.1 GFWV01020600 MAA45328.1 CH933808 EDW10569.2 KRG05321.1 KRG05322.1 KRG05323.1 CH480838 EDW49306.1 OUUW01000001 SPP72666.1 CH940648 EDW60165.2 CM000071 EAL25727.3 KA645248 KA649138 AFP59877.1 AFP63767.1 GAKP01019660 GAKP01019658 JAC39292.1 GANP01015996 JAB68472.1 GEFM01004575 GEGO01005300 JAP71221.1 JAR90104.1 GDHF01018958 GDHF01017444 GDHF01016739 GDHF01014475 GDHF01014324 JAI33356.1 JAI34870.1 JAI35575.1 JAI37839.1 JAI37990.1 SPP72667.1 CH902619 EDV37263.2 JXJN01013915 AFH08270.1 CH479183 EDW36157.1 CH963849 EDW74057.1 JRES01000628 KNC29796.1 CH916375 EDV98465.1 AMQN01011584 KB308963 ELT95899.1
KQ459585 KPI98196.1 KQ971338 EFA01541.2 GEDC01012082 JAS25216.1 GEZM01045043 JAV77955.1 ABLF02018593 ABLF02018595 ABLF02019204 ABLF02019206 ABLF02019207 GGMS01007349 MBY76552.1 GEDC01001238 JAS36060.1 GEDC01031480 GEDC01004466 JAS05818.1 JAS32832.1 GEDC01011793 JAS25505.1 GECZ01024932 JAS44837.1 AAAB01008849 GECU01025788 GECU01021631 JAS81918.1 JAS86075.1 GDIP01048315 LRGB01001569 JAM55400.1 KZS11735.1 GDIP01055915 JAM47800.1 GECU01005087 JAT02620.1 GL732619 EFX70577.1 GDIQ01028417 JAN66320.1 GECZ01019278 JAS50491.1 GDIP01182642 JAJ40760.1 GGFM01006004 MBW26755.1 GGFJ01004195 MBW53336.1 EAL41047.3 GAPW01001045 JAC12553.1 GGFL01000253 MBW64431.1 GGFL01000254 MBW64432.1 GGFL01000252 MBW64430.1 GGFK01004713 MBW38034.1 GGFK01004729 MBW38050.1 JH431832 CH477865 EAT35498.1 GDIP01145996 JAJ77406.1 AXCN02002284 APCN01005116 KA648118 AFP62747.1 LNIX01000006 OXA52717.1 GFJQ02000677 JAW06293.1 CM002911 KMY96524.1 KMY96521.1 KMY96522.1 KMY96523.1 AE013599 AY069169 M97204 GFJQ02008307 JAV98662.1 CH954179 EDV57121.1 GBXI01002933 JAD11359.1 CM000158 EDW92650.1 KRK00624.1 GFWV01020600 MAA45328.1 CH933808 EDW10569.2 KRG05321.1 KRG05322.1 KRG05323.1 CH480838 EDW49306.1 OUUW01000001 SPP72666.1 CH940648 EDW60165.2 CM000071 EAL25727.3 KA645248 KA649138 AFP59877.1 AFP63767.1 GAKP01019660 GAKP01019658 JAC39292.1 GANP01015996 JAB68472.1 GEFM01004575 GEGO01005300 JAP71221.1 JAR90104.1 GDHF01018958 GDHF01017444 GDHF01016739 GDHF01014475 GDHF01014324 JAI33356.1 JAI34870.1 JAI35575.1 JAI37839.1 JAI37990.1 SPP72667.1 CH902619 EDV37263.2 JXJN01013915 AFH08270.1 CH479183 EDW36157.1 CH963849 EDW74057.1 JRES01000628 KNC29796.1 CH916375 EDV98465.1 AMQN01011584 KB308963 ELT95899.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000053268
UP000007266
UP000007819
+ More
UP000075903 UP000076858 UP000069272 UP000000305 UP000007062 UP000008820 UP000075886 UP000075840 UP000075885 UP000095301 UP000198287 UP000075881 UP000000803 UP000095300 UP000075900 UP000008711 UP000002282 UP000075920 UP000009192 UP000001292 UP000076408 UP000192221 UP000268350 UP000008792 UP000001819 UP000079169 UP000007801 UP000092460 UP000091820 UP000008744 UP000007798 UP000037069 UP000001070 UP000014760
UP000075903 UP000076858 UP000069272 UP000000305 UP000007062 UP000008820 UP000075886 UP000075840 UP000075885 UP000095301 UP000198287 UP000075881 UP000000803 UP000095300 UP000075900 UP000008711 UP000002282 UP000075920 UP000009192 UP000001292 UP000076408 UP000192221 UP000268350 UP000008792 UP000001819 UP000079169 UP000007801 UP000092460 UP000091820 UP000008744 UP000007798 UP000037069 UP000001070 UP000014760
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A3S2M767
A0A212EH97
A0A194RC27
A0A194PXX5
D2A1E8
A0A1B6DHR0
+ More
A0A1Y1M0X6 J9K827 A0A2S2QHA3 A0A1B6EDU2 A0A1B6E4H6 A0A1B6DIP4 A0A1B6F3T8 A0A1S4FVT7 A0A182V8R7 A0A453Z0L3 A0A1B6IGP3 A0A0P5Z6D6 A0A0P5YP19 A0A182F458 A0A1B6JTW9 E9HC82 A0A0P6GWM5 A0A1B6FJY8 A0A0P5BXF3 A0A2M3ZE25 A0A2M4BJT6 Q5TUG4 A0A023ETC3 A0A2M4CGZ8 A0A2M4CGF9 A0A2M4CGG0 A0A2M4AB69 A0A2M4ABB7 T1J3Y4 Q16MD5 A0A0P5EA85 A0A1Y9H9H6 A0A453Z0X3 A0A182HX16 A0A182PSD2 T1PJK6 A0A1I8MG78 A0A226E6X8 A0A182KGY3 A0A1Z5LFW6 A0A0J9RM83 A0A0J9RKN1 Q06003 A0A1Z5KU94 A0A1I8NSU6 A0A182R910 B3NRA2 A0A0A1XJ46 B4PBZ6 A0A293MZG4 A0A182W877 Q06003-4 B4KSS0 B4IHD0 A0A182YE34 A0A1W4V5W9 A0A3B0JHF5 B4LNU5 Q28Z87 T1PB96 A0A034VB16 V5H2L8 A0A131Y031 A0A0K8V8G7 A0A3Q0JKC6 A0A3B0IY81 B3MCD9 A0A1B0BGP2 Q06003-3 A0A0B4K8B7 A0A1A9WV64 B4GI37 B4MPL8 A0A0L0CC15 B4JVJ7 R7TQG9
A0A1Y1M0X6 J9K827 A0A2S2QHA3 A0A1B6EDU2 A0A1B6E4H6 A0A1B6DIP4 A0A1B6F3T8 A0A1S4FVT7 A0A182V8R7 A0A453Z0L3 A0A1B6IGP3 A0A0P5Z6D6 A0A0P5YP19 A0A182F458 A0A1B6JTW9 E9HC82 A0A0P6GWM5 A0A1B6FJY8 A0A0P5BXF3 A0A2M3ZE25 A0A2M4BJT6 Q5TUG4 A0A023ETC3 A0A2M4CGZ8 A0A2M4CGF9 A0A2M4CGG0 A0A2M4AB69 A0A2M4ABB7 T1J3Y4 Q16MD5 A0A0P5EA85 A0A1Y9H9H6 A0A453Z0X3 A0A182HX16 A0A182PSD2 T1PJK6 A0A1I8MG78 A0A226E6X8 A0A182KGY3 A0A1Z5LFW6 A0A0J9RM83 A0A0J9RKN1 Q06003 A0A1Z5KU94 A0A1I8NSU6 A0A182R910 B3NRA2 A0A0A1XJ46 B4PBZ6 A0A293MZG4 A0A182W877 Q06003-4 B4KSS0 B4IHD0 A0A182YE34 A0A1W4V5W9 A0A3B0JHF5 B4LNU5 Q28Z87 T1PB96 A0A034VB16 V5H2L8 A0A131Y031 A0A0K8V8G7 A0A3Q0JKC6 A0A3B0IY81 B3MCD9 A0A1B0BGP2 Q06003-3 A0A0B4K8B7 A0A1A9WV64 B4GI37 B4MPL8 A0A0L0CC15 B4JVJ7 R7TQG9
PDB
5ZC4
E-value=2.5972e-10,
Score=157
Ontologies
GO
Topology
Subcellular location
Membrane
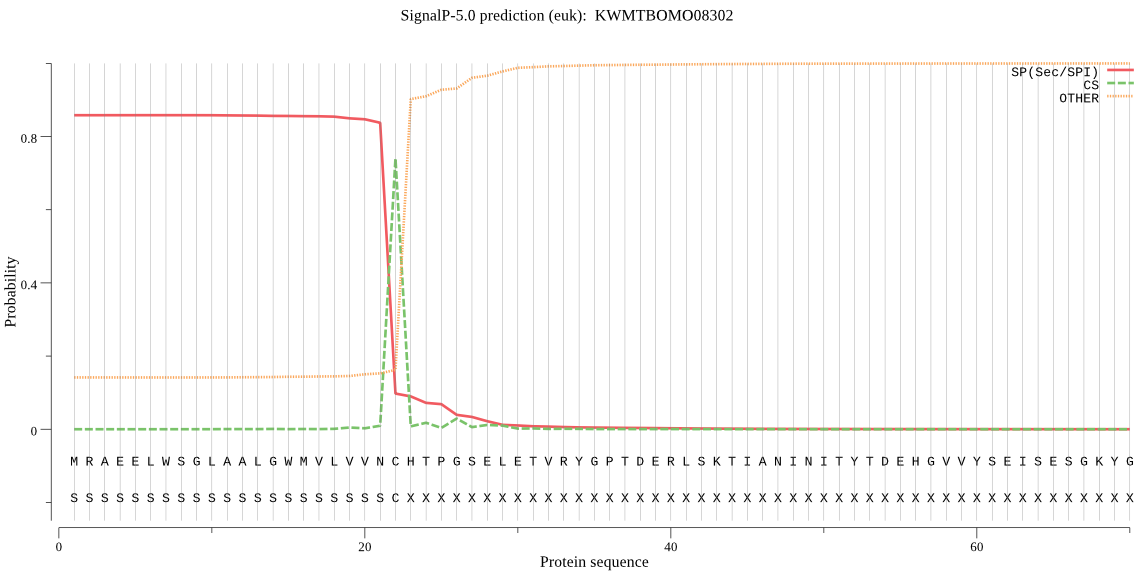
SignalP
Position: 1 - 22,
Likelihood: 0.858832
Length:
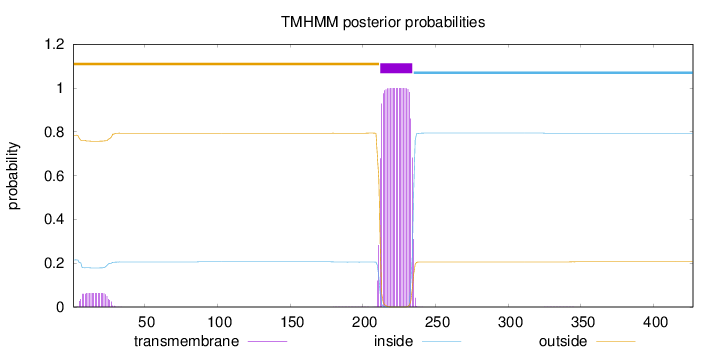
427
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.17819
Exp number, first 60 AAs:
1.28651
Total prob of N-in:
0.21510
outside
1 - 211
TMhelix
212 - 234
inside
235 - 427
Population Genetic Test Statistics
Pi
233.094998
Theta
165.345046
Tajima's D
1.199478
CLR
0.259305
CSRT
0.714414279286036
Interpretation
Uncertain
