Pre Gene Modal
BGIBMGA009534
Annotation
PREDICTED:_cytoskeleton-associated_protein_5_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.617 Mitochondrial Reliability : 1.055
Sequence
CDS
ATGCTAGTGGCGTCGGGCGGGCGGCTGCTGGGCGGCGTGGCGGCGGGGCTGCGGGCCCGCTTCCAGCCCTACGCCGCGCCCTGCGTGCAGGCCATCCTCGAGAAGTTTAAAGAGAAGAAGCCGAACGTTGTCGCGGCGCTGCGGGAGGCCATCGACGCCATCTACCCTTCCACCAATCTGGAAGTAATAATGGAAGACATGCTAGCTGCGTTTGACAATAAGAACCCGTCTATAAAGGCCGAGAGCGCCCTGTTCTTGGCTCGCGCGCTGTGCCACACTCAGCCTGCCGCTTTTAATAAGAAACTCTTAAAGGCCTATGTGGCAGGCTTATTAAAACTATTGGAGAGCGCTGATCCAGCTGTAAGAGACGCGGCTGCAGAAGCTCTAGGCACGGCCACTAAACTGGTGGGCGAGAAGAACATAGCACCGCATATTGGAAAGTTGGACAACCTGAAGGAACAGAAGATCAAGGAATTTGCAGATAAGGCTGTTATTCAAGTAAAACTAACAGCGCCAAAAGAAGCTAAACCAAAATCAGCTCCTGTTGCAAAAGACACCAAGACAGCTGGTTCCTCCCAGCCCAAACTTGTTAAGCGACCAGCCTCAGCCGTAAAAAAGAGCAGTGCTGCCACAAAGAAGCCCGTAGCAGGACGAGGTCAGATCAGTCCTCCGAGGGAACAAGAACTGTCTTCTGAAGAAATAGACGCACGCGCTGAACAACTACTTAACCCTGATGTTATTAGTGGTCTTGGAGACAGCAATTGGAAGACTCGTCTGTCAGCTGTCCAGTCTTATTACTCTACAGTACAAAGCTTATCTACAAACGACGCCACAGCCCAAGTACTGTATCGTATATTGAACAAAAAACCAGGTCTCAAAGACACTAATGTACAGGTTCTCAAAGCAAGATTAGAAGTATGCAAGTACATTACAGAGAATTTTCCGATTTCCACAACTGCCGCAGATTTCATTCTCCAAGATGTAGTCACGAAATTGGGCGAGAGTTCCTGCTCTGCTCTGTCAGCCGATTGTCTGACCTCCCTGGCGGAGTCGTGCGGCCTCGAACATGTCTTAAACGAAGGATTGACTTTCGCTATGGACTCTCAGAAGAATCCCAAAGTTCAGTCCGAAATATTCAACTGGATGTCTAATGCAATCAAAGAGTTTGGATTCAATATGAACATAAAGTCGATAGTTGCGCACTCGAAGAAGGCTCTGTCGGCCACGAACCCCAATGTCCGCACTTCGGTAGTGAACTTCCTGGGAGTGTTGTCGCTGTACGTGGGCCCCGCGCTCATCGGACACTTCGACAGCGAGAAGCCGGCCACCAAGCAGCTCATCAGCCTGGAGCTCGACAAGTACGCCAACAGCACGCCTCCGACCCCCACCAGGCAGGTCGGCAGTGTCACCAAGTCCGCCAGCAAAGTGTCGATGGGCGAGGACCTAGAAGAAGTATACGAAGACGCGACTGAGGACGAGCCGGCCCCCGCCGAGGACTCGCGGCCGCGGACGGACATCGCGCCCCTCATCACGGAGGCGCTTGTGGCCGAAATGAATGATAAGAACTGGAAGGTTCGTAACGAAGCTTTAGACAAAGTGAAGGCAATAATAACGAATTCATCGCCAATAAAACCGAGTCTGGGCGAGTTGCCGCCGGCCCTGGTCGCCCGCCTGCTGGACTCCAACTCCAAACTAGCACAGACCGCTCTCAATATATGCGAGGCGCTGGCCACAGCGATGGGCCCTAAGTGCAAACAGCACGTAAGGACATTCTTCCCGGCATTCTTTCAAGCCCTGGGCGACAACAAGCAGTGGATCCGCGCGGCGGCGGTGTCGTGCGTGGAGGCGTGGTGCCGCGAGGCCGGGCCCGCCGCGCCGCTCGACGGGGAGATGCTGTTCGACGCGCTGAAGGCGGGCTCGCCTGTACTCAGGGCCTCGCTGCTGCAGTGGCTCGCTGAACACCTGCCTAATGTACCTGCCAAATCATTTCCAAGAGAAGAGCTCTCAGTGTGTGTGGGGACGCTATTCGCGTGTATAGAGGACCGGGCGGCGGACGTGCGGAAAGCCGCAGGGGACTGCGTTTTCCCGTTCATGTTGCATTTGGGATACGAAGCGATGCACAAGCAGCTTGAAAAATTGAAGCCCGGCTCCAAAACAGTAGTTCTGGCAGCATTAGACAAAGCTCGTCCAAATTTGCCAATTCAACCGTTACCAACCAAAACCAAGAAGGAGGAGACCAAGGGAGTTAAGACCGGAGGGGCAATGAAGAAAGAAGCAGAAAAGAAAAGCGCCCCTGTAGGTAAAGGAAAGGTGGTTAAACCGTCGTCGGCATCAAGTCGCGGTAAGAAAGACGATGAGGATTGTTCCCCGCTATTACCGAACAACAATGCCAAGAATCAACGTATTATTGACGACCAGAAACTAAAAGTATTGAAATGGAATTTCACAACACCACGAGAGGAGTTCTTCGAGCTACTCAAAGATCAAATGGGCACCGCCGGTCTCAATAAACAACTTATCGCCAACATGTTTCATAGTGATTTCAGATATCATCTAAAAGCGATCGAAGCATTATCTGAAGATCTTCACGAGAATGGTCCAGCGTTAGTTTCCAACTTGGACCTGGTCCTCAAGTGGCTGACTCTGCGATTTTTCGATACAAATCCATCAGTCATACTCAAGGGACTCGAGTACTTGTCTATGGTATTTCACCATCTCATAGATTGCGACTACACAATGGCGGAATATGAAGCAAATTGTTTTATTCCTTATTTGGTGTTAAAGGTGGGGGATCCAAAAGATACAGTGCGCAACGGAGTCAAAGCACTCTTCAGACAAATATGCGTTGTGTATTCTGTTACAAAATTGTTTGGGTACCTCATGGAGGGACTCAAATCTAAGAATGCGAGGCAAAGAGCCGGTGAGCACATATTATATTATAATAACTTATAA
Protein
MLVASGGRLLGGVAAGLRARFQPYAAPCVQAILEKFKEKKPNVVAALREAIDAIYPSTNLEVIMEDMLAAFDNKNPSIKAESALFLARALCHTQPAAFNKKLLKAYVAGLLKLLESADPAVRDAAAEALGTATKLVGEKNIAPHIGKLDNLKEQKIKEFADKAVIQVKLTAPKEAKPKSAPVAKDTKTAGSSQPKLVKRPASAVKKSSAATKKPVAGRGQISPPREQELSSEEIDARAEQLLNPDVISGLGDSNWKTRLSAVQSYYSTVQSLSTNDATAQVLYRILNKKPGLKDTNVQVLKARLEVCKYITENFPISTTAADFILQDVVTKLGESSCSALSADCLTSLAESCGLEHVLNEGLTFAMDSQKNPKVQSEIFNWMSNAIKEFGFNMNIKSIVAHSKKALSATNPNVRTSVVNFLGVLSLYVGPALIGHFDSEKPATKQLISLELDKYANSTPPTPTRQVGSVTKSASKVSMGEDLEEVYEDATEDEPAPAEDSRPRTDIAPLITEALVAEMNDKNWKVRNEALDKVKAIITNSSPIKPSLGELPPALVARLLDSNSKLAQTALNICEALATAMGPKCKQHVRTFFPAFFQALGDNKQWIRAAAVSCVEAWCREAGPAAPLDGEMLFDALKAGSPVLRASLLQWLAEHLPNVPAKSFPREELSVCVGTLFACIEDRAADVRKAAGDCVFPFMLHLGYEAMHKQLEKLKPGSKTVVLAALDKARPNLPIQPLPTKTKKEETKGVKTGGAMKKEAEKKSAPVGKGKVVKPSSASSRGKKDDEDCSPLLPNNNAKNQRIIDDQKLKVLKWNFTTPREEFFELLKDQMGTAGLNKQLIANMFHSDFRYHLKAIEALSEDLHENGPALVSNLDLVLKWLTLRFFDTNPSVILKGLEYLSMVFHHLIDCDYTMAEYEANCFIPYLVLKVGDPKDTVRNGVKALFRQICVVYSVTKLFGYLMEGLKSKNARQRAGEHILYYNNL
Summary
Uniprot
H9JJ33
A0A3S2NK60
A0A2H1V9Y1
A0A194PXY3
A0A212EPW6
A0A2H1VPJ7
+ More
F4WJ83 A0A158NED1 A0A336MCP1 A0A336KTU9 E2C892 A0A195BX28 A0A3L8E1Z2 A0A195FL51 A0A151WVX7 A0A151J6L3 A0A232F9C1 E2B1J0 A0A195CAH0 K7IX50 A0A0J7L889 A0A1Y1JZC9 A0A1Y1JZU4 A0A1Y1K4W3 A0A0C9QAW6 A0A2A3E4Z3 A0A1J1IJA6 V9IC27 A0A088A8Y4 A0A1W4X2D8 A0A1B0CAC3 W4VRR1 A0A154PK94 A0A1B6CWX2 A0A1B6C9W5 A0A1B6DJ97 A0A1B6CVQ0 A0A182GZY0 Q0IFD6 A0A1S4FBN3 A0A067QZY8 A0A1Q3F0V9 A0A1Q3F0T5 A0A1Q3F0R6 A0A1Q3F0Z5 A0A1Q3F179 B0WL70 A0A0Q9X519 A0A0Q9WV40 A0A0Q9WVJ9 B4NL16 A0A3B0JN84 A0A3B0KD72 A0A3B0JN73 A0A3B0JRN5 A0A2J7R1V0 A0A2J7R1U2 A0A2J7R1U0 W8APC6 W8APC2 W8B9B2 A0A0M4F2L5 W8AZP4 W8ADX9 A0A0Q9XBE3 A0A0Q9X968 A0A0Q9X2I2 A0A1B6ME06 B4KAF5 A0A1B6KZK6 A0A0Q9XD54 A0A0Q9X1B7 A0A0Q9XCU6 A0A0P6DCK9 A0A182MWU9 A0A0P5M3G3 A0A0P6HDW5 A0A0P5JD27 A0A0P6HZD3 A0A0P5QYL8 A0A0P5WUY2 A0A0P6CN56 A0A0N8C405 A0A0P4Y8G8 A0A0N8BEZ6 A0A0P5YXS8 A0A0P5TXU0 A0A0L0BP62 A0A0P5QZA1 A0A0P5SE51 A0A0P8XLD2 A0A0N8NZX8 A0A0P5R9N3 Q7QHF0 A0A0P8Y8N7 A0A0P8XLD8 A0A0P9ABF2 A0A0P8XLH5 A0A1W4VED1 A0A1W4VE16
F4WJ83 A0A158NED1 A0A336MCP1 A0A336KTU9 E2C892 A0A195BX28 A0A3L8E1Z2 A0A195FL51 A0A151WVX7 A0A151J6L3 A0A232F9C1 E2B1J0 A0A195CAH0 K7IX50 A0A0J7L889 A0A1Y1JZC9 A0A1Y1JZU4 A0A1Y1K4W3 A0A0C9QAW6 A0A2A3E4Z3 A0A1J1IJA6 V9IC27 A0A088A8Y4 A0A1W4X2D8 A0A1B0CAC3 W4VRR1 A0A154PK94 A0A1B6CWX2 A0A1B6C9W5 A0A1B6DJ97 A0A1B6CVQ0 A0A182GZY0 Q0IFD6 A0A1S4FBN3 A0A067QZY8 A0A1Q3F0V9 A0A1Q3F0T5 A0A1Q3F0R6 A0A1Q3F0Z5 A0A1Q3F179 B0WL70 A0A0Q9X519 A0A0Q9WV40 A0A0Q9WVJ9 B4NL16 A0A3B0JN84 A0A3B0KD72 A0A3B0JN73 A0A3B0JRN5 A0A2J7R1V0 A0A2J7R1U2 A0A2J7R1U0 W8APC6 W8APC2 W8B9B2 A0A0M4F2L5 W8AZP4 W8ADX9 A0A0Q9XBE3 A0A0Q9X968 A0A0Q9X2I2 A0A1B6ME06 B4KAF5 A0A1B6KZK6 A0A0Q9XD54 A0A0Q9X1B7 A0A0Q9XCU6 A0A0P6DCK9 A0A182MWU9 A0A0P5M3G3 A0A0P6HDW5 A0A0P5JD27 A0A0P6HZD3 A0A0P5QYL8 A0A0P5WUY2 A0A0P6CN56 A0A0N8C405 A0A0P4Y8G8 A0A0N8BEZ6 A0A0P5YXS8 A0A0P5TXU0 A0A0L0BP62 A0A0P5QZA1 A0A0P5SE51 A0A0P8XLD2 A0A0N8NZX8 A0A0P5R9N3 Q7QHF0 A0A0P8Y8N7 A0A0P8XLD8 A0A0P9ABF2 A0A0P8XLH5 A0A1W4VED1 A0A1W4VE16
Pubmed
EMBL
BABH01040275
BABH01040276
BABH01040277
BABH01040278
BABH01040279
RSAL01000016
+ More
RVE53041.1 ODYU01001448 SOQ37660.1 KQ459585 KPI98192.1 AGBW02013389 OWR43501.1 ODYU01003678 SOQ42750.1 GL888182 EGI65661.1 ADTU01013252 UFQS01000939 UFQT01000939 SSX07860.1 SSX28094.1 SSX07861.1 SSX28095.1 GL453586 EFN75833.1 KQ976401 KYM92523.1 QOIP01000001 RLU26463.1 KQ981490 KYN41076.1 KQ982696 KYQ51976.1 KQ979851 KYN18848.1 NNAY01000601 OXU27451.1 GL444934 EFN60447.1 KQ978023 KYM97869.1 AAZX01000600 AAZX01021298 LBMM01000231 KMR03598.1 GEZM01096672 JAV54684.1 GEZM01096673 JAV54683.1 GEZM01096671 JAV54685.1 GBYB01000369 JAG70136.1 KZ288379 PBC26564.1 CVRI01000049 CRK99142.1 JR039300 AEY58625.1 AJWK01003648 AJWK01003649 GANO01000598 JAB59273.1 KQ434943 KZC12217.1 GEDC01019384 GEDC01013928 JAS17914.1 JAS23370.1 GEDC01027020 GEDC01016294 JAS10278.1 JAS21004.1 GEDC01014783 GEDC01011593 JAS22515.1 JAS25705.1 GEDC01019893 GEDC01015728 JAS17405.1 JAS21570.1 JXUM01100352 JXUM01100353 KQ564562 KXJ72084.1 CH477356 EAT42779.1 KK853147 KDR10714.1 GFDL01013871 JAV21174.1 GFDL01013883 JAV21162.1 GFDL01013892 JAV21153.1 GFDL01013887 JAV21158.1 GFDL01013807 JAV21238.1 DS231980 EDS30243.1 CH964272 KRF99997.1 KRF99998.1 KRF99996.1 EDW84219.1 OUUW01000008 SPP83675.1 SPP83676.1 SPP83674.1 SPP83673.1 NEVH01008202 PNF34802.1 PNF34801.1 PNF34799.1 GAMC01020192 JAB86363.1 GAMC01020197 JAB86358.1 GAMC01020196 JAB86359.1 CP012526 ALC45345.1 GAMC01020194 JAB86361.1 GAMC01020195 JAB86360.1 CH933806 KRG01671.1 KRG01668.1 KRG01670.1 KRG01673.1 GEBQ01005816 JAT34161.1 EDW15668.1 GEBQ01023081 JAT16896.1 KRG01667.1 KRG01669.1 KRG01672.1 GDIQ01079174 JAN15563.1 AXCM01001359 GDIQ01170041 JAK81684.1 GDIQ01033599 JAN61138.1 GDIQ01203684 GDIQ01203683 GDIQ01203682 JAK48041.1 GDIQ01012946 JAN81791.1 GDIQ01117018 GDIQ01117017 JAL34708.1 GDIP01081324 JAM22391.1 GDIP01012114 JAM91601.1 GDIQ01117019 JAL34707.1 GDIP01234856 JAI88545.1 GDIQ01184291 GDIQ01184290 GDIQ01184289 GDIQ01184288 JAK67434.1 GDIP01053084 JAM50631.1 GDIP01121065 JAL82649.1 JRES01001582 KNC21768.1 GDIQ01108118 JAL43608.1 GDIQ01105717 JAL46009.1 CH902629 KPU75560.1 KPU75564.1 GDIQ01105716 JAL46010.1 AAAB01008816 EAA05144.4 KPU75562.1 KPU75558.1 KPU75559.1 KPU75561.1
RVE53041.1 ODYU01001448 SOQ37660.1 KQ459585 KPI98192.1 AGBW02013389 OWR43501.1 ODYU01003678 SOQ42750.1 GL888182 EGI65661.1 ADTU01013252 UFQS01000939 UFQT01000939 SSX07860.1 SSX28094.1 SSX07861.1 SSX28095.1 GL453586 EFN75833.1 KQ976401 KYM92523.1 QOIP01000001 RLU26463.1 KQ981490 KYN41076.1 KQ982696 KYQ51976.1 KQ979851 KYN18848.1 NNAY01000601 OXU27451.1 GL444934 EFN60447.1 KQ978023 KYM97869.1 AAZX01000600 AAZX01021298 LBMM01000231 KMR03598.1 GEZM01096672 JAV54684.1 GEZM01096673 JAV54683.1 GEZM01096671 JAV54685.1 GBYB01000369 JAG70136.1 KZ288379 PBC26564.1 CVRI01000049 CRK99142.1 JR039300 AEY58625.1 AJWK01003648 AJWK01003649 GANO01000598 JAB59273.1 KQ434943 KZC12217.1 GEDC01019384 GEDC01013928 JAS17914.1 JAS23370.1 GEDC01027020 GEDC01016294 JAS10278.1 JAS21004.1 GEDC01014783 GEDC01011593 JAS22515.1 JAS25705.1 GEDC01019893 GEDC01015728 JAS17405.1 JAS21570.1 JXUM01100352 JXUM01100353 KQ564562 KXJ72084.1 CH477356 EAT42779.1 KK853147 KDR10714.1 GFDL01013871 JAV21174.1 GFDL01013883 JAV21162.1 GFDL01013892 JAV21153.1 GFDL01013887 JAV21158.1 GFDL01013807 JAV21238.1 DS231980 EDS30243.1 CH964272 KRF99997.1 KRF99998.1 KRF99996.1 EDW84219.1 OUUW01000008 SPP83675.1 SPP83676.1 SPP83674.1 SPP83673.1 NEVH01008202 PNF34802.1 PNF34801.1 PNF34799.1 GAMC01020192 JAB86363.1 GAMC01020197 JAB86358.1 GAMC01020196 JAB86359.1 CP012526 ALC45345.1 GAMC01020194 JAB86361.1 GAMC01020195 JAB86360.1 CH933806 KRG01671.1 KRG01668.1 KRG01670.1 KRG01673.1 GEBQ01005816 JAT34161.1 EDW15668.1 GEBQ01023081 JAT16896.1 KRG01667.1 KRG01669.1 KRG01672.1 GDIQ01079174 JAN15563.1 AXCM01001359 GDIQ01170041 JAK81684.1 GDIQ01033599 JAN61138.1 GDIQ01203684 GDIQ01203683 GDIQ01203682 JAK48041.1 GDIQ01012946 JAN81791.1 GDIQ01117018 GDIQ01117017 JAL34708.1 GDIP01081324 JAM22391.1 GDIP01012114 JAM91601.1 GDIQ01117019 JAL34707.1 GDIP01234856 JAI88545.1 GDIQ01184291 GDIQ01184290 GDIQ01184289 GDIQ01184288 JAK67434.1 GDIP01053084 JAM50631.1 GDIP01121065 JAL82649.1 JRES01001582 KNC21768.1 GDIQ01108118 JAL43608.1 GDIQ01105717 JAL46009.1 CH902629 KPU75560.1 KPU75564.1 GDIQ01105716 JAL46010.1 AAAB01008816 EAA05144.4 KPU75562.1 KPU75558.1 KPU75559.1 KPU75561.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000007755
UP000005205
+ More
UP000008237 UP000078540 UP000279307 UP000078541 UP000075809 UP000078492 UP000215335 UP000000311 UP000078542 UP000002358 UP000036403 UP000242457 UP000183832 UP000005203 UP000192223 UP000092461 UP000076502 UP000069940 UP000249989 UP000008820 UP000027135 UP000002320 UP000007798 UP000268350 UP000235965 UP000092553 UP000009192 UP000075883 UP000037069 UP000007801 UP000007062 UP000192221
UP000008237 UP000078540 UP000279307 UP000078541 UP000075809 UP000078492 UP000215335 UP000000311 UP000078542 UP000002358 UP000036403 UP000242457 UP000183832 UP000005203 UP000192223 UP000092461 UP000076502 UP000069940 UP000249989 UP000008820 UP000027135 UP000002320 UP000007798 UP000268350 UP000235965 UP000092553 UP000009192 UP000075883 UP000037069 UP000007801 UP000007062 UP000192221
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JJ33
A0A3S2NK60
A0A2H1V9Y1
A0A194PXY3
A0A212EPW6
A0A2H1VPJ7
+ More
F4WJ83 A0A158NED1 A0A336MCP1 A0A336KTU9 E2C892 A0A195BX28 A0A3L8E1Z2 A0A195FL51 A0A151WVX7 A0A151J6L3 A0A232F9C1 E2B1J0 A0A195CAH0 K7IX50 A0A0J7L889 A0A1Y1JZC9 A0A1Y1JZU4 A0A1Y1K4W3 A0A0C9QAW6 A0A2A3E4Z3 A0A1J1IJA6 V9IC27 A0A088A8Y4 A0A1W4X2D8 A0A1B0CAC3 W4VRR1 A0A154PK94 A0A1B6CWX2 A0A1B6C9W5 A0A1B6DJ97 A0A1B6CVQ0 A0A182GZY0 Q0IFD6 A0A1S4FBN3 A0A067QZY8 A0A1Q3F0V9 A0A1Q3F0T5 A0A1Q3F0R6 A0A1Q3F0Z5 A0A1Q3F179 B0WL70 A0A0Q9X519 A0A0Q9WV40 A0A0Q9WVJ9 B4NL16 A0A3B0JN84 A0A3B0KD72 A0A3B0JN73 A0A3B0JRN5 A0A2J7R1V0 A0A2J7R1U2 A0A2J7R1U0 W8APC6 W8APC2 W8B9B2 A0A0M4F2L5 W8AZP4 W8ADX9 A0A0Q9XBE3 A0A0Q9X968 A0A0Q9X2I2 A0A1B6ME06 B4KAF5 A0A1B6KZK6 A0A0Q9XD54 A0A0Q9X1B7 A0A0Q9XCU6 A0A0P6DCK9 A0A182MWU9 A0A0P5M3G3 A0A0P6HDW5 A0A0P5JD27 A0A0P6HZD3 A0A0P5QYL8 A0A0P5WUY2 A0A0P6CN56 A0A0N8C405 A0A0P4Y8G8 A0A0N8BEZ6 A0A0P5YXS8 A0A0P5TXU0 A0A0L0BP62 A0A0P5QZA1 A0A0P5SE51 A0A0P8XLD2 A0A0N8NZX8 A0A0P5R9N3 Q7QHF0 A0A0P8Y8N7 A0A0P8XLD8 A0A0P9ABF2 A0A0P8XLH5 A0A1W4VED1 A0A1W4VE16
F4WJ83 A0A158NED1 A0A336MCP1 A0A336KTU9 E2C892 A0A195BX28 A0A3L8E1Z2 A0A195FL51 A0A151WVX7 A0A151J6L3 A0A232F9C1 E2B1J0 A0A195CAH0 K7IX50 A0A0J7L889 A0A1Y1JZC9 A0A1Y1JZU4 A0A1Y1K4W3 A0A0C9QAW6 A0A2A3E4Z3 A0A1J1IJA6 V9IC27 A0A088A8Y4 A0A1W4X2D8 A0A1B0CAC3 W4VRR1 A0A154PK94 A0A1B6CWX2 A0A1B6C9W5 A0A1B6DJ97 A0A1B6CVQ0 A0A182GZY0 Q0IFD6 A0A1S4FBN3 A0A067QZY8 A0A1Q3F0V9 A0A1Q3F0T5 A0A1Q3F0R6 A0A1Q3F0Z5 A0A1Q3F179 B0WL70 A0A0Q9X519 A0A0Q9WV40 A0A0Q9WVJ9 B4NL16 A0A3B0JN84 A0A3B0KD72 A0A3B0JN73 A0A3B0JRN5 A0A2J7R1V0 A0A2J7R1U2 A0A2J7R1U0 W8APC6 W8APC2 W8B9B2 A0A0M4F2L5 W8AZP4 W8ADX9 A0A0Q9XBE3 A0A0Q9X968 A0A0Q9X2I2 A0A1B6ME06 B4KAF5 A0A1B6KZK6 A0A0Q9XD54 A0A0Q9X1B7 A0A0Q9XCU6 A0A0P6DCK9 A0A182MWU9 A0A0P5M3G3 A0A0P6HDW5 A0A0P5JD27 A0A0P6HZD3 A0A0P5QYL8 A0A0P5WUY2 A0A0P6CN56 A0A0N8C405 A0A0P4Y8G8 A0A0N8BEZ6 A0A0P5YXS8 A0A0P5TXU0 A0A0L0BP62 A0A0P5QZA1 A0A0P5SE51 A0A0P8XLD2 A0A0N8NZX8 A0A0P5R9N3 Q7QHF0 A0A0P8Y8N7 A0A0P8XLD8 A0A0P9ABF2 A0A0P8XLH5 A0A1W4VED1 A0A1W4VE16
PDB
5VJC
E-value=1.83493e-73,
Score=705
Ontologies
KEGG
GO
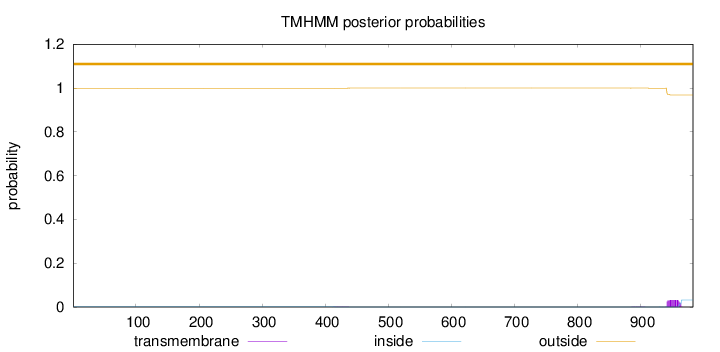
Topology
Length:
983
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.69744
Exp number, first 60 AAs:
0.00151
Total prob of N-in:
0.00090
outside
1 - 983
Population Genetic Test Statistics
Pi
20.237035
Theta
19.335313
Tajima's D
0.213926
CLR
158.083376
CSRT
0.44002799860007
Interpretation
Uncertain
