Gene
KWMTBOMO08292
Pre Gene Modal
BGIBMGA009538
Annotation
PREDICTED:_uncharacterized_protein_LOC105841802_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.822 PlasmaMembrane Reliability : 1.987
Sequence
CDS
ATGTTCCAGCCTATATTCACCAACTACGGACTCTGTTTCGCATTCAACAGCCTACCCCTGAACGGAATGACGAACAGCACACTGGCTTGGCAAAAATCTTTTAATCCACAATCGGCAGCGAAATCATTACTGTGGGGCTTAGATATCGGTTATCCGAAGACATTTCCGCCAGACGTCAATATGCAACCACTGAGGGTAATGGCATCGGGAGAGGCTAACGGCTTGGGTGTAGAACTGTTTTTAAATACAAGCGAACATCAATATGCTTGCGATGGAAATAGCTTGGGTTTCACGGTCTTGATAAGCTCTCCAGCTGACCACGTCTACACCAGCACCGTACTCAGAATCCCAATGGACAGAATGACCACAATCGAAGTGTCGCCAATAACCTACAAGACCGATTCATCTTTAAGGTCACTAGCCCCGTATCAGAGGCAGTGCTCCTTTCAGAACGAGAGAAGACTTGAGCATTACGAATATTACACTGACAGTAACTGTCAGCATGATTTGTTGGTGAAGGAGGCTCGGAGGATGTGTAACTGTGTACTCTTCAACTGGCCAAGAGTAAGTCTGCAGGAATCAGTCTGTTCGACTGATGAGGATTTCCAATGCATTACCAATGTAAACGGGAAAGTTGAGGAACAATTAATTTACGCTTATTACGCTGACAGCGAAGCACGGAGGGAGCAACGGGAACAATCCAAATCTTGTCATCCTTCTTGCAATGATGTGATCTACTCCAGCCAGGTGTTCTACTCAGACCTTATTAGAGACTACGGCGATGGTACACCTCATTGGGGAATTCCGGGCCAAAGTGAAGTAACCCAGATCAACGTGCACTTTTACGAAGACTTCTTCCTGGGCCAACACCGCCACGCACAATACGACGATTATTACTTTGCAGGTGCTATTGGGGGACTGTTGAGTCTATTCTTGGGCTTTAGCATCATCAGTGTCGCCGAACTTGTGTACTTTGTAATCTTACGTCCTGCTTATACTATTTTAAAGGAAATGTATTATCAAAGAAATAAATAA
Protein
MFQPIFTNYGLCFAFNSLPLNGMTNSTLAWQKSFNPQSAAKSLLWGLDIGYPKTFPPDVNMQPLRVMASGEANGLGVELFLNTSEHQYACDGNSLGFTVLISSPADHVYTSTVLRIPMDRMTTIEVSPITYKTDSSLRSLAPYQRQCSFQNERRLEHYEYYTDSNCQHDLLVKEARRMCNCVLFNWPRVSLQESVCSTDEDFQCITNVNGKVEEQLIYAYYADSEARREQREQSKSCHPSCNDVIYSSQVFYSDLIRDYGDGTPHWGIPGQSEVTQINVHFYEDFFLGQHRHAQYDDYYFAGAIGGLLSLFLGFSIISVAELVYFVILRPAYTILKEMYYQRNK
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
A0A2H1WMH5
A0A2H1X2I1
A0A3S2M7V0
A0A194PXW7
A0A0L7L3Y1
A0A194R870
+ More
A0A0L0CPB4 B4N9B6 B5RJJ1 O46342 B4QVK8 A0A182QS19 A0A0Q9WPD7 A0A182W972 W8C7R3 A0A182PBQ5 A0A3B0K6B3 B4PUF8 U4UGG3 Q17DP0 B3M0N3 A0A1S4F6X7 A0A182MLZ3 Q7Q066 A0A1I8N7J9 A0A182HQ19 B4M371 A0A1W4UQ48 A0A1W4V2C9 A0A182KSP4 D2A5Z0 A0A0M3QZM1 B4GFB0 A0A182THH2 Q296D6 A0A182MYI9 A0A0B4K691 B4JJ58 A0A1I8N4A3 A0A1I8N2I2 B3P232 A0A182XD77 A0A182UXK6 A0A1B6HF37 A0A182K171 A0A0K8VEQ2 A0A0A1WE01 A0A182Y1U0 T1H7R9 A0A084WHX7 A0A034WIR7 A0A1B0CKI0 Q17MX5 A0A1I8NWY6 A0A1I8NWY9 N6U8E9 A0A182R5F2 A0A1S4EX71 B3NFC4 B0WG99 A0A1B0GH50 T1GJ31 A0A0J9RQW9 B4HV75 B4L493 A0A1I8M339 Q9VS73 A0A1I8Q523 A0A182GRG8 A0A182G5B3 A0A034WQE2 A0A1J1HMY9 A0A1L8DFH2 A0A1B0D0X6 A0A1A9X593 A0A1L8DF46 A0A1J1HSC5 B0X0R9 A0A1I8M8B2 A0A139WKH1 D6WXM6 B4I3R4 A0A1B0GL64 A0A0M3QW07 A0A1I8PU93 A0A1I8Q8M8 A0A1I8N2I9 A0A3B0JWU3 A0A182H7C8 B4ML83 A0A1L8DF10 B4PC79 A0A182RAM9 Q2M133 A0A1I8Q5L9 B4GUR0 Q17MX3 B3M5Z2 B0W821
A0A0L0CPB4 B4N9B6 B5RJJ1 O46342 B4QVK8 A0A182QS19 A0A0Q9WPD7 A0A182W972 W8C7R3 A0A182PBQ5 A0A3B0K6B3 B4PUF8 U4UGG3 Q17DP0 B3M0N3 A0A1S4F6X7 A0A182MLZ3 Q7Q066 A0A1I8N7J9 A0A182HQ19 B4M371 A0A1W4UQ48 A0A1W4V2C9 A0A182KSP4 D2A5Z0 A0A0M3QZM1 B4GFB0 A0A182THH2 Q296D6 A0A182MYI9 A0A0B4K691 B4JJ58 A0A1I8N4A3 A0A1I8N2I2 B3P232 A0A182XD77 A0A182UXK6 A0A1B6HF37 A0A182K171 A0A0K8VEQ2 A0A0A1WE01 A0A182Y1U0 T1H7R9 A0A084WHX7 A0A034WIR7 A0A1B0CKI0 Q17MX5 A0A1I8NWY6 A0A1I8NWY9 N6U8E9 A0A182R5F2 A0A1S4EX71 B3NFC4 B0WG99 A0A1B0GH50 T1GJ31 A0A0J9RQW9 B4HV75 B4L493 A0A1I8M339 Q9VS73 A0A1I8Q523 A0A182GRG8 A0A182G5B3 A0A034WQE2 A0A1J1HMY9 A0A1L8DFH2 A0A1B0D0X6 A0A1A9X593 A0A1L8DF46 A0A1J1HSC5 B0X0R9 A0A1I8M8B2 A0A139WKH1 D6WXM6 B4I3R4 A0A1B0GL64 A0A0M3QW07 A0A1I8PU93 A0A1I8Q8M8 A0A1I8N2I9 A0A3B0JWU3 A0A182H7C8 B4ML83 A0A1L8DF10 B4PC79 A0A182RAM9 Q2M133 A0A1I8Q5L9 B4GUR0 Q17MX3 B3M5Z2 B0W821
Pubmed
26354079
26227816
26108605
17994087
9600094
9545267
+ More
9425162 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 17550304 23537049 17510324 12364791 14747013 17210077 25315136 20966253 18362917 19820115 15632085 25830018 25244985 24438588 25348373 22936249 26483478
9425162 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 17550304 23537049 17510324 12364791 14747013 17210077 25315136 20966253 18362917 19820115 15632085 25830018 25244985 24438588 25348373 22936249 26483478
EMBL
ODYU01009675
SOQ54269.1
ODYU01012968
SOQ59531.1
RSAL01000016
RVE53035.1
+ More
KQ459585 KPI98186.1 JTDY01003182 KOB70009.1 KQ460627 KPJ13455.1 JRES01000091 KNC34208.1 CH964232 EDW80549.1 BT044465 ACH92530.1 AF043264 AE014297 Y16240 AAC38824.1 AAF52108.2 CAA76129.1 CM000364 EDX11633.1 AXCN02001041 CH933810 KRF94123.1 GAMC01000377 JAC06179.1 OUUW01000007 SPP83610.1 CM000160 EDW95680.2 KB632186 ERL89696.1 CH477292 EAT44535.1 CH902617 EDV44280.2 AXCM01007075 AAAB01008986 EAA00234.4 APCN01003288 CH940651 EDW65246.1 KQ971345 EFA05007.2 CP012528 ALC49619.1 CH479182 EDW34295.1 CM000070 EAL28522.3 AFH06228.1 CH916370 EDV99610.1 CH954181 EDV47805.1 GECU01034485 JAS73221.1 GDHF01014958 JAI37356.1 GBXI01017063 JAC97228.1 ACPB03005214 ATLV01023896 KE525347 KFB49821.1 GAKP01003476 JAC55476.1 AJWK01016308 CH477203 EAT48027.1 APGK01038999 APGK01039000 KB740966 ENN76926.1 CH954178 EDV50466.2 DS231923 EDS26795.1 AJWK01003969 AJWK01003970 AJWK01003971 AJWK01003972 CAQQ02088033 CM002912 KMY98102.1 CH480817 EDW50846.1 EDW07371.2 AE014296 AAF50555.3 JXUM01082415 KQ563311 KXJ74083.1 JXUM01043285 KQ561357 KXJ78848.1 GAKP01001166 JAC57786.1 CVRI01000004 CRK87601.1 GFDF01009000 JAV05084.1 AJVK01010113 GFDF01009001 JAV05083.1 CVRI01000018 CRK90292.1 DS232244 EDS38311.1 KQ971327 KYB28426.1 KQ971362 EFA08873.2 CH480821 EDW54857.1 AJWK01034404 AJWK01034405 CP012525 ALC43319.1 OUUW01000002 SPP77191.1 JXUM01116018 KQ565940 KXJ70514.1 CH963847 EDW73141.1 GFDF01009117 JAV04967.1 CM000159 EDW93764.1 CH379069 EAL30743.2 CH479191 EDW26343.1 EAT48029.1 CH902618 EDV40708.1 DS231856 EDS38483.1
KQ459585 KPI98186.1 JTDY01003182 KOB70009.1 KQ460627 KPJ13455.1 JRES01000091 KNC34208.1 CH964232 EDW80549.1 BT044465 ACH92530.1 AF043264 AE014297 Y16240 AAC38824.1 AAF52108.2 CAA76129.1 CM000364 EDX11633.1 AXCN02001041 CH933810 KRF94123.1 GAMC01000377 JAC06179.1 OUUW01000007 SPP83610.1 CM000160 EDW95680.2 KB632186 ERL89696.1 CH477292 EAT44535.1 CH902617 EDV44280.2 AXCM01007075 AAAB01008986 EAA00234.4 APCN01003288 CH940651 EDW65246.1 KQ971345 EFA05007.2 CP012528 ALC49619.1 CH479182 EDW34295.1 CM000070 EAL28522.3 AFH06228.1 CH916370 EDV99610.1 CH954181 EDV47805.1 GECU01034485 JAS73221.1 GDHF01014958 JAI37356.1 GBXI01017063 JAC97228.1 ACPB03005214 ATLV01023896 KE525347 KFB49821.1 GAKP01003476 JAC55476.1 AJWK01016308 CH477203 EAT48027.1 APGK01038999 APGK01039000 KB740966 ENN76926.1 CH954178 EDV50466.2 DS231923 EDS26795.1 AJWK01003969 AJWK01003970 AJWK01003971 AJWK01003972 CAQQ02088033 CM002912 KMY98102.1 CH480817 EDW50846.1 EDW07371.2 AE014296 AAF50555.3 JXUM01082415 KQ563311 KXJ74083.1 JXUM01043285 KQ561357 KXJ78848.1 GAKP01001166 JAC57786.1 CVRI01000004 CRK87601.1 GFDF01009000 JAV05084.1 AJVK01010113 GFDF01009001 JAV05083.1 CVRI01000018 CRK90292.1 DS232244 EDS38311.1 KQ971327 KYB28426.1 KQ971362 EFA08873.2 CH480821 EDW54857.1 AJWK01034404 AJWK01034405 CP012525 ALC43319.1 OUUW01000002 SPP77191.1 JXUM01116018 KQ565940 KXJ70514.1 CH963847 EDW73141.1 GFDF01009117 JAV04967.1 CM000159 EDW93764.1 CH379069 EAL30743.2 CH479191 EDW26343.1 EAT48029.1 CH902618 EDV40708.1 DS231856 EDS38483.1
Proteomes
UP000283053
UP000053268
UP000037510
UP000053240
UP000037069
UP000007798
+ More
UP000000803 UP000000304 UP000075886 UP000009192 UP000075920 UP000075885 UP000268350 UP000002282 UP000030742 UP000008820 UP000007801 UP000075883 UP000007062 UP000095301 UP000075840 UP000008792 UP000192221 UP000075882 UP000007266 UP000092553 UP000008744 UP000075902 UP000001819 UP000075884 UP000001070 UP000008711 UP000076407 UP000075903 UP000075881 UP000076408 UP000015103 UP000030765 UP000092461 UP000095300 UP000019118 UP000075900 UP000002320 UP000015102 UP000001292 UP000069940 UP000249989 UP000183832 UP000092462 UP000091820
UP000000803 UP000000304 UP000075886 UP000009192 UP000075920 UP000075885 UP000268350 UP000002282 UP000030742 UP000008820 UP000007801 UP000075883 UP000007062 UP000095301 UP000075840 UP000008792 UP000192221 UP000075882 UP000007266 UP000092553 UP000008744 UP000075902 UP000001819 UP000075884 UP000001070 UP000008711 UP000076407 UP000075903 UP000075881 UP000076408 UP000015103 UP000030765 UP000092461 UP000095300 UP000019118 UP000075900 UP000002320 UP000015102 UP000001292 UP000069940 UP000249989 UP000183832 UP000092462 UP000091820
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1WMH5
A0A2H1X2I1
A0A3S2M7V0
A0A194PXW7
A0A0L7L3Y1
A0A194R870
+ More
A0A0L0CPB4 B4N9B6 B5RJJ1 O46342 B4QVK8 A0A182QS19 A0A0Q9WPD7 A0A182W972 W8C7R3 A0A182PBQ5 A0A3B0K6B3 B4PUF8 U4UGG3 Q17DP0 B3M0N3 A0A1S4F6X7 A0A182MLZ3 Q7Q066 A0A1I8N7J9 A0A182HQ19 B4M371 A0A1W4UQ48 A0A1W4V2C9 A0A182KSP4 D2A5Z0 A0A0M3QZM1 B4GFB0 A0A182THH2 Q296D6 A0A182MYI9 A0A0B4K691 B4JJ58 A0A1I8N4A3 A0A1I8N2I2 B3P232 A0A182XD77 A0A182UXK6 A0A1B6HF37 A0A182K171 A0A0K8VEQ2 A0A0A1WE01 A0A182Y1U0 T1H7R9 A0A084WHX7 A0A034WIR7 A0A1B0CKI0 Q17MX5 A0A1I8NWY6 A0A1I8NWY9 N6U8E9 A0A182R5F2 A0A1S4EX71 B3NFC4 B0WG99 A0A1B0GH50 T1GJ31 A0A0J9RQW9 B4HV75 B4L493 A0A1I8M339 Q9VS73 A0A1I8Q523 A0A182GRG8 A0A182G5B3 A0A034WQE2 A0A1J1HMY9 A0A1L8DFH2 A0A1B0D0X6 A0A1A9X593 A0A1L8DF46 A0A1J1HSC5 B0X0R9 A0A1I8M8B2 A0A139WKH1 D6WXM6 B4I3R4 A0A1B0GL64 A0A0M3QW07 A0A1I8PU93 A0A1I8Q8M8 A0A1I8N2I9 A0A3B0JWU3 A0A182H7C8 B4ML83 A0A1L8DF10 B4PC79 A0A182RAM9 Q2M133 A0A1I8Q5L9 B4GUR0 Q17MX3 B3M5Z2 B0W821
A0A0L0CPB4 B4N9B6 B5RJJ1 O46342 B4QVK8 A0A182QS19 A0A0Q9WPD7 A0A182W972 W8C7R3 A0A182PBQ5 A0A3B0K6B3 B4PUF8 U4UGG3 Q17DP0 B3M0N3 A0A1S4F6X7 A0A182MLZ3 Q7Q066 A0A1I8N7J9 A0A182HQ19 B4M371 A0A1W4UQ48 A0A1W4V2C9 A0A182KSP4 D2A5Z0 A0A0M3QZM1 B4GFB0 A0A182THH2 Q296D6 A0A182MYI9 A0A0B4K691 B4JJ58 A0A1I8N4A3 A0A1I8N2I2 B3P232 A0A182XD77 A0A182UXK6 A0A1B6HF37 A0A182K171 A0A0K8VEQ2 A0A0A1WE01 A0A182Y1U0 T1H7R9 A0A084WHX7 A0A034WIR7 A0A1B0CKI0 Q17MX5 A0A1I8NWY6 A0A1I8NWY9 N6U8E9 A0A182R5F2 A0A1S4EX71 B3NFC4 B0WG99 A0A1B0GH50 T1GJ31 A0A0J9RQW9 B4HV75 B4L493 A0A1I8M339 Q9VS73 A0A1I8Q523 A0A182GRG8 A0A182G5B3 A0A034WQE2 A0A1J1HMY9 A0A1L8DFH2 A0A1B0D0X6 A0A1A9X593 A0A1L8DF46 A0A1J1HSC5 B0X0R9 A0A1I8M8B2 A0A139WKH1 D6WXM6 B4I3R4 A0A1B0GL64 A0A0M3QW07 A0A1I8PU93 A0A1I8Q8M8 A0A1I8N2I9 A0A3B0JWU3 A0A182H7C8 B4ML83 A0A1L8DF10 B4PC79 A0A182RAM9 Q2M133 A0A1I8Q5L9 B4GUR0 Q17MX3 B3M5Z2 B0W821
Ontologies
GO
PANTHER
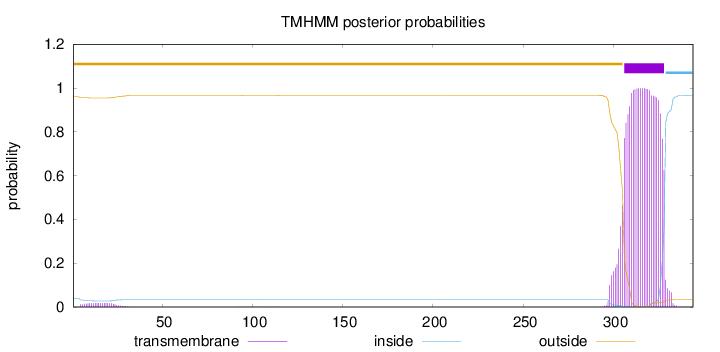
Topology
Length:
344
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.07873
Exp number, first 60 AAs:
0.35758
Total prob of N-in:
0.03941
outside
1 - 305
TMhelix
306 - 328
inside
329 - 344
Population Genetic Test Statistics
Pi
341.315505
Theta
208.198134
Tajima's D
1.702541
CLR
0.15046
CSRT
0.827708614569272
Interpretation
Uncertain
