Gene
KWMTBOMO08291
Pre Gene Modal
BGIBMGA009539
Annotation
PREDICTED:_uncharacterized_protein_LOC105841802_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.969
Sequence
CDS
ATGGGTTTAGCTATTTGTTTATTACTTGAATGCAGAATTTCAGATATATCTTCGTGGCGTTGGATTCTTGTCGATCTAGCGAAGCGTCTCGAAGAATCCATCGAGGCTCTGCAACATGAGCACAACGCTCTGCGAGTAGTGATCCGGAGAATCGATGATGAACTGAACATCTACTCCAGACAGGGTGCGCTGACGACACTGGGTGCTTTGGCTGATAGCGTTGAAGATGCTATAATTCAGGAACAAAATTTTCTACGTAACGAAAAAAATAAATTTGAAAAGATGTTAACAGAATTAGAAAAGCAAATAATTAATGTAGAGAAAACGAAAAAGCGAATAGAATTGGATATTCTGAAGAAACAGCAAACTTTAGACATAGAAGATGCTTGTGCCAACATTGACTCTGGTTCAATATTGACGAATAGAAAGAAAAAAAAACGGAGAAGCTCGCCATTATCTCGTTGGGAAAACAGGTGTGATTCATTAAAAAAAGCTGGTCTTAAATCTTTAAGAAATGCCATAATAACAAGACAGCAGGTTAGAGGTGCGAGAATTCAAATTTCAATAGCTGCCCAGGGCTATGCCGCTAAGGTGGACTCGATTCTCAAAAGGCGACTGCACGCTAATCAACTGAAATTTCAAGATCTATGTTGGCAGAGAGAAGAGGCCATTAAAGACTTCAATGTTTTGGAAGAGGAATTAACGACGACGGAACAGAATCTTCTAGAAAACTTAGAACAAGAAAGACTTATTGAAACCAGAATAACTGACCGCTCTCAGAGACCCGTCGGAGAGCTCACCAAAGATGACGTGGATAGAAAATTGAGAGATGAACTTGGCAGATTAAGGAATTTCACTAACGAATTAAGAAAAAACCGGTCCAGAATCACCACTCTTCAAAATCACATAACAAACTGTATAAGTCGCATTGAGTGTTGCTCCGAAGATCTCAACCAAGTAATCAATTTGGATGAAGAGAGGATGAAGTTAAGAAACGGAGAAATCCAAGGAGAACCATCAGAAGAGAATGTCAAGGGTATTCAGCCTTCTGGAACCATAGCGAAAAGGTCTCAAATGAGATCGAACGAAATATTGACTGCGATTACCGAAGAGGACGAAGACGATTATCCATTTAATGACTAA
Protein
MGLAICLLLECRISDISSWRWILVDLAKRLEESIEALQHEHNALRVVIRRIDDELNIYSRQGALTTLGALADSVEDAIIQEQNFLRNEKNKFEKMLTELEKQIINVEKTKKRIELDILKKQQTLDIEDACANIDSGSILTNRKKKKRRSSPLSRWENRCDSLKKAGLKSLRNAIITRQQVRGARIQISIAAQGYAAKVDSILKRRLHANQLKFQDLCWQREEAIKDFNVLEEELTTTEQNLLENLEQERLIETRITDRSQRPVGELTKDDVDRKLRDELGRLRNFTNELRKNRSRITTLQNHITNCISRIECCSEDLNQVINLDEERMKLRNGEIQGEPSEENVKGIQPSGTIAKRSQMRSNEILTAITEEDEDDYPFND
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
H9JJ38
A0A3S2TRC6
A0A2H1X2I1
A0A2A4JA11
A0A194R7G1
A0A194Q449
+ More
A0A212EU56 A0A0L7L9S9 E2AZR8 F4WY84 A0A195AWS9 A0A158NHR5 A0A3M0IS71 A0A0J7L5D8 A0A195EHE9 A0A026WWJ2 A0A3L8D8X4 A0A151X6Y6 A0A195CB88 A0A2A4J8U6 A0A212F0U9 A0A2H1W8M1 A0A195FES6 A0A3B5QJU3 A0A437BWG4 A0A336LSN8 Q0IFA2 D6WGQ5 A0A182GIL1 A0A182H7Y6 A0A3B3UM87 A0A3B5KZS8 A0A087XMN0 A0A3B4DN22 A0A3B3Y6I2 A0A0M9A8X8 A0A3B3CSJ2 A0A0L7KYV2 A0A194QAF5 A0A226PEM6 A0A0N1II92 A0A3B3CR54 A0A315V0N9
A0A212EU56 A0A0L7L9S9 E2AZR8 F4WY84 A0A195AWS9 A0A158NHR5 A0A3M0IS71 A0A0J7L5D8 A0A195EHE9 A0A026WWJ2 A0A3L8D8X4 A0A151X6Y6 A0A195CB88 A0A2A4J8U6 A0A212F0U9 A0A2H1W8M1 A0A195FES6 A0A3B5QJU3 A0A437BWG4 A0A336LSN8 Q0IFA2 D6WGQ5 A0A182GIL1 A0A182H7Y6 A0A3B3UM87 A0A3B5KZS8 A0A087XMN0 A0A3B4DN22 A0A3B3Y6I2 A0A0M9A8X8 A0A3B3CSJ2 A0A0L7KYV2 A0A194QAF5 A0A226PEM6 A0A0N1II92 A0A3B3CR54 A0A315V0N9
Pubmed
EMBL
BABH01040265
RSAL01000016
RVE53034.1
ODYU01012968
SOQ59531.1
NWSH01002456
+ More
PCG68262.1 KQ460627 KPJ13454.1 KQ459585 KPI98185.1 AGBW02012462 OWR45018.1 JTDY01002146 KOB72006.1 GL444277 EFN61047.1 GL888439 EGI60841.1 KQ976725 KYM76626.1 ADTU01016031 ADTU01016032 QRBI01000238 RMB91248.1 LBMM01000686 KMQ97763.1 KQ978957 KYN27297.1 KK107078 EZA60445.1 QOIP01000011 RLU16761.1 KQ982476 KYQ56070.1 KQ978023 KYM98067.1 NWSH01002602 PCG67880.1 AGBW02011024 OWR47360.1 ODYU01007032 SOQ49418.1 KQ981636 KYN38866.1 RSAL01000004 RVE54601.1 UFQT01000102 SSX20011.1 CH477389 EAT41984.1 KQ971327 EEZ99614.1 JXUM01066478 KQ562400 KXJ75996.1 JXUM01117605 KQ566100 KXJ70398.1 AYCK01008130 KQ435724 KOX78353.1 JTDY01004374 KOB68231.1 KQ459249 KPJ02414.1 AWGT02000094 OXB77847.1 KQ460324 KPJ15851.1 NHOQ01002476 PWA16763.1
PCG68262.1 KQ460627 KPJ13454.1 KQ459585 KPI98185.1 AGBW02012462 OWR45018.1 JTDY01002146 KOB72006.1 GL444277 EFN61047.1 GL888439 EGI60841.1 KQ976725 KYM76626.1 ADTU01016031 ADTU01016032 QRBI01000238 RMB91248.1 LBMM01000686 KMQ97763.1 KQ978957 KYN27297.1 KK107078 EZA60445.1 QOIP01000011 RLU16761.1 KQ982476 KYQ56070.1 KQ978023 KYM98067.1 NWSH01002602 PCG67880.1 AGBW02011024 OWR47360.1 ODYU01007032 SOQ49418.1 KQ981636 KYN38866.1 RSAL01000004 RVE54601.1 UFQT01000102 SSX20011.1 CH477389 EAT41984.1 KQ971327 EEZ99614.1 JXUM01066478 KQ562400 KXJ75996.1 JXUM01117605 KQ566100 KXJ70398.1 AYCK01008130 KQ435724 KOX78353.1 JTDY01004374 KOB68231.1 KQ459249 KPJ02414.1 AWGT02000094 OXB77847.1 KQ460324 KPJ15851.1 NHOQ01002476 PWA16763.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000000311 UP000007755 UP000078540 UP000005205 UP000269221 UP000036403 UP000078492 UP000053097 UP000279307 UP000075809 UP000078542 UP000078541 UP000002852 UP000008820 UP000007266 UP000069940 UP000249989 UP000261500 UP000261380 UP000028760 UP000261440 UP000261480 UP000053105 UP000261560 UP000198419
UP000037510 UP000000311 UP000007755 UP000078540 UP000005205 UP000269221 UP000036403 UP000078492 UP000053097 UP000279307 UP000075809 UP000078542 UP000078541 UP000002852 UP000008820 UP000007266 UP000069940 UP000249989 UP000261500 UP000261380 UP000028760 UP000261440 UP000261480 UP000053105 UP000261560 UP000198419
PRIDE
Pfam
Interpro
IPR000435
Tektin
+ More
IPR001873 ENaC
IPR036705 Ribosyl_crysJ1_sf
IPR032474 Argonaute_N
IPR012337 RNaseH-like_sf
IPR032473 Argonaute_Mid_dom
IPR005502 Ribosyl_crysJ1
IPR036085 PAZ_dom_sf
IPR036397 RNaseH_sf
IPR014811 ArgoL1
IPR032472 ArgoL2
IPR003100 PAZ_dom
IPR003165 Piwi
IPR006825 Eclosion
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR001873 ENaC
IPR036705 Ribosyl_crysJ1_sf
IPR032474 Argonaute_N
IPR012337 RNaseH-like_sf
IPR032473 Argonaute_Mid_dom
IPR005502 Ribosyl_crysJ1
IPR036085 PAZ_dom_sf
IPR036397 RNaseH_sf
IPR014811 ArgoL1
IPR032472 ArgoL2
IPR003100 PAZ_dom
IPR003165 Piwi
IPR006825 Eclosion
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
Gene 3D
ProteinModelPortal
H9JJ38
A0A3S2TRC6
A0A2H1X2I1
A0A2A4JA11
A0A194R7G1
A0A194Q449
+ More
A0A212EU56 A0A0L7L9S9 E2AZR8 F4WY84 A0A195AWS9 A0A158NHR5 A0A3M0IS71 A0A0J7L5D8 A0A195EHE9 A0A026WWJ2 A0A3L8D8X4 A0A151X6Y6 A0A195CB88 A0A2A4J8U6 A0A212F0U9 A0A2H1W8M1 A0A195FES6 A0A3B5QJU3 A0A437BWG4 A0A336LSN8 Q0IFA2 D6WGQ5 A0A182GIL1 A0A182H7Y6 A0A3B3UM87 A0A3B5KZS8 A0A087XMN0 A0A3B4DN22 A0A3B3Y6I2 A0A0M9A8X8 A0A3B3CSJ2 A0A0L7KYV2 A0A194QAF5 A0A226PEM6 A0A0N1II92 A0A3B3CR54 A0A315V0N9
A0A212EU56 A0A0L7L9S9 E2AZR8 F4WY84 A0A195AWS9 A0A158NHR5 A0A3M0IS71 A0A0J7L5D8 A0A195EHE9 A0A026WWJ2 A0A3L8D8X4 A0A151X6Y6 A0A195CB88 A0A2A4J8U6 A0A212F0U9 A0A2H1W8M1 A0A195FES6 A0A3B5QJU3 A0A437BWG4 A0A336LSN8 Q0IFA2 D6WGQ5 A0A182GIL1 A0A182H7Y6 A0A3B3UM87 A0A3B5KZS8 A0A087XMN0 A0A3B4DN22 A0A3B3Y6I2 A0A0M9A8X8 A0A3B3CSJ2 A0A0L7KYV2 A0A194QAF5 A0A226PEM6 A0A0N1II92 A0A3B3CR54 A0A315V0N9
Ontologies
GO
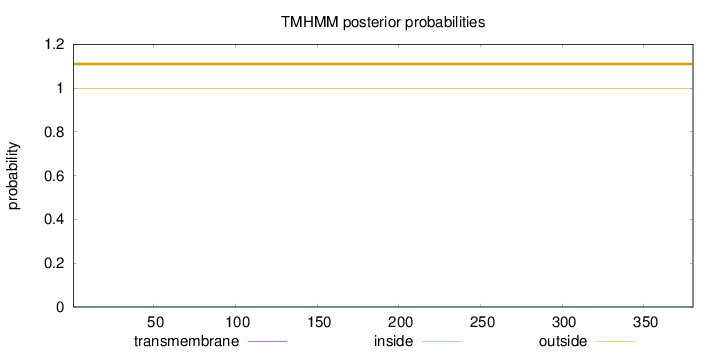
Topology
Length:
380
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000860000000000001
Exp number, first 60 AAs:
0.00065
Total prob of N-in:
0.00088
outside
1 - 380
Population Genetic Test Statistics
Pi
292.712265
Theta
191.137177
Tajima's D
1.571028
CLR
0.06164
CSRT
0.807859607019649
Interpretation
Uncertain
