Gene
KWMTBOMO08289
Pre Gene Modal
BGIBMGA009540
Annotation
PREDICTED:_tetratricopeptide_repeat_protein_5-like_[Papilio_xuthus]
Full name
Tetratricopeptide repeat protein 5
Alternative Name
Stress-responsive activator of p300
Location in the cell
Cytoplasmic Reliability : 2.757
Sequence
CDS
ATGTCGGAATCAGAAGTGGAAGCTGATATAATAGAAAATGTTGCAGAAGTCCTGGATAATCTATCGAAAGAACTAGAAGAATTGTATAGGTTTCGAAACTTGTTTTTTGAAAATCAACCAATTAGTATGGCTGCAGAAAAAAATAAGCAAGTTGAGGAAAAGAAAAATCTACTCCTAGAAAAGTTTGAATCCATAGATGATGCACAAATACCATCAGCATCGAGAGCCCAGTTCTTGTACCTTAAAGGACGTTGCTATAATATTAGTCCAACATATGATGCAAGAGCTACACATTGTCTCAGCAAAGCAGTAAAGTGGAATCCTCACATGGTGGATGCTTGGAATGAACTTGGCGAATGTTATTGGAAGAATATGAATGTTAAAGAGGCTAAAGCCAGCTTCGAAGCTGCTTTGAAACATGAAAGGAATCGACTCTCACTACGTTGTCTGTCAATCATTTTGAGACAAGAAGGTGGTGACAGAAAACGTCCTGATTCCACCAATGCGATTGGCACAAGTGTTGAACTTGCAAAAGAGGCTGTATCTCAGGACACAAAAGACGGTATCTCATGGACAGTATTAGGGAATGCTTTATTATGCCAGTTCTTTATGGTCTCCCAAGATCCAGCAATTCTGAAATTATGCATGACAGCATATAGGCAAGCCTGGGCTGATCCTGTTGCTAAGGGGCAGCCGGATTTATACTACAATAAGGCTATTGCCTTGAAATATGACGAAATATACGATGAAGCATTAGAAAACTTTGAATATGCGTGTCGTTTGGATCCCCCGTGGGAACCACCGCAGTTGGAACTCCAAAAGTTAAAACAATATTTAAACGAAACAAATGAATTAGTACGAACTAAAGGAAAGATTAAAGTTAAGAAACTGACACAGATGGTACAGTCGATAGACGAGAAGATGCTAGGAATATACAGCCCAGAAGGTTTGCACACATTCGGTGGTCGTCGCAACGTGACCCTAGAGCTGTGTCGGTTGGAAGATTTGCACGAGGGACTCAATGAAGGAAAAGTCATATTAGGAAGAGTCGTTGGCTCCATTCACAATGAAAATGCTGTGCCTTTTTCGTTCGCAATCGTTGGTCAGGGCATGCAGTGCGCCTGCGTCACCGTCTACAACTGGGCCTCGGGTCGGGGAGCCATCATTGGAGACTGCGTGACTATACCGGAACCACTACTGACCGTACACAAACACGCATCCGAATTGGCCAGCTACGAGTTCAAGTCGATCCGCGTCAACAACCCGACGCTGCTGCTGGTGAACGGCAGGCGCGTCGGCCGCGAGCAGTACGCGAGCACGCGCGTCACCAGCACCTACGAGCTGCACTGA
Protein
MSESEVEADIIENVAEVLDNLSKELEELYRFRNLFFENQPISMAAEKNKQVEEKKNLLLEKFESIDDAQIPSASRAQFLYLKGRCYNISPTYDARATHCLSKAVKWNPHMVDAWNELGECYWKNMNVKEAKASFEAALKHERNRLSLRCLSIILRQEGGDRKRPDSTNAIGTSVELAKEAVSQDTKDGISWTVLGNALLCQFFMVSQDPAILKLCMTAYRQAWADPVAKGQPDLYYNKAIALKYDEIYDEALENFEYACRLDPPWEPPQLELQKLKQYLNETNELVRTKGKIKVKKLTQMVQSIDEKMLGIYSPEGLHTFGGRRNVTLELCRLEDLHEGLNEGKVILGRVVGSIHNENAVPFSFAIVGQGMQCACVTVYNWASGRGAIIGDCVTIPEPLLTVHKHASELASYEFKSIRVNNPTLLLVNGRRVGREQYASTRVTSTYELH
Summary
Description
Adapter protein involved in p53/TP53 response that acts by regulating and mediating the assembly of multi-protein complexes. Required to facilitate the interaction between JMY and p300/EP300 and increase p53/TP53-dependent transcription and apoptosis. Prevents p53/TP53 degradation by MDM2 (By similarity).
Subunit
Interacts with JMY and p300/EP300.
Keywords
3D-structure
Complete proteome
Cytoplasm
DNA damage
DNA repair
Nucleus
Phosphoprotein
Polymorphism
Reference proteome
Repeat
TPR repeat
Feature
chain Tetratricopeptide repeat protein 5
sequence variant In dbSNP:rs34675160.
sequence variant In dbSNP:rs34675160.
Uniprot
H9JJ39
A0A194PXX8
A0A194RC17
A0A2A4J8R8
A0A3S2P5N7
A0A0C9RYX1
+ More
A0A1Y1M5P0 E9J0S8 A0A232ESW6 K7J6R9 A0A151WPS4 A0A151JXG6 A0A195ECV5 A0A158NXR6 A0A154PE41 A0A195AZQ6 A0A026WKS7 F4WTH1 E2BAA0 A0A2J7R321 A0A087ZRS6 E1ZYT4 A0A0L7R5W3 D1ZZR8 A0A195CKV9 A0A1B6HXA3 A0A1Y1MB57 A0A0M9A450 E0VHW1 A0A1B6F4K9 A0A2J7R335 A0A1B6DEG0 A0A2A3EP51 A0A067RBV7 A0A310SWV8 A7RRB4 A0A1B6KVA8 A0A3L8E224 A0A0B7AF55 A0A2P8ZDS4 K1PNB1 A0A3M6U3G2 C3XU94 V4B2K3 T1J2A1 A0A3Q0JKL5 A0A1S3JSL4 A0A1S3JT74 A0A0B7AEN7 A0A1Y1M944 A0A3Q0E0Q4 G1LG62 A0A1W2W544 A0A3Q7VBL8 A0A3Q0K5Y5 A0A384BH51 A0A452RHI0 A0A2K6U289 F6Q337 U6CTX2 G3SQJ0 U3C0A8 F7D0C6 A0A2R9BP76 Q8N0Z6 G3QDN9 M3W130 A0A2I3RW55 G3WGM9 H2NKH1 A0A2K5QJJ6 A0A0B7AE47 S7PHZ4 G1PS04 G5B5Q3 A0A0D9RW71 A0A2K5F1G4 A0A2I3H703 A0JMT7 Q6NRA9 A0A383ZZM7 I3MEN8 A0A2Y9EAG2 A0A2U3WUI0 L5MB24 A0A3S1BJB2 A0A2K5YA58 A0A3Q7RI40 A0A250YIA0 L5K6D8 A0A091CWR2 K9IJW8 M3Z354 A0A2K5M0H4 M7BDF6 H0VDM7 F6WBF3
A0A1Y1M5P0 E9J0S8 A0A232ESW6 K7J6R9 A0A151WPS4 A0A151JXG6 A0A195ECV5 A0A158NXR6 A0A154PE41 A0A195AZQ6 A0A026WKS7 F4WTH1 E2BAA0 A0A2J7R321 A0A087ZRS6 E1ZYT4 A0A0L7R5W3 D1ZZR8 A0A195CKV9 A0A1B6HXA3 A0A1Y1MB57 A0A0M9A450 E0VHW1 A0A1B6F4K9 A0A2J7R335 A0A1B6DEG0 A0A2A3EP51 A0A067RBV7 A0A310SWV8 A7RRB4 A0A1B6KVA8 A0A3L8E224 A0A0B7AF55 A0A2P8ZDS4 K1PNB1 A0A3M6U3G2 C3XU94 V4B2K3 T1J2A1 A0A3Q0JKL5 A0A1S3JSL4 A0A1S3JT74 A0A0B7AEN7 A0A1Y1M944 A0A3Q0E0Q4 G1LG62 A0A1W2W544 A0A3Q7VBL8 A0A3Q0K5Y5 A0A384BH51 A0A452RHI0 A0A2K6U289 F6Q337 U6CTX2 G3SQJ0 U3C0A8 F7D0C6 A0A2R9BP76 Q8N0Z6 G3QDN9 M3W130 A0A2I3RW55 G3WGM9 H2NKH1 A0A2K5QJJ6 A0A0B7AE47 S7PHZ4 G1PS04 G5B5Q3 A0A0D9RW71 A0A2K5F1G4 A0A2I3H703 A0JMT7 Q6NRA9 A0A383ZZM7 I3MEN8 A0A2Y9EAG2 A0A2U3WUI0 L5MB24 A0A3S1BJB2 A0A2K5YA58 A0A3Q7RI40 A0A250YIA0 L5K6D8 A0A091CWR2 K9IJW8 M3Z354 A0A2K5M0H4 M7BDF6 H0VDM7 F6WBF3
Pubmed
19121390
26354079
28004739
21282665
28648823
20075255
+ More
21347285 24508170 21719571 20798317 18362917 19820115 20566863 24845553 17615350 30249741 29403074 22992520 30382153 18563158 23254933 20010809 19892987 25243066 22722832 14702039 12508121 15489334 21269460 22362889 22398555 17975172 16136131 21709235 21993624 21993625 26763976 27762356 28087693 23258410 23624526 17495919
21347285 24508170 21719571 20798317 18362917 19820115 20566863 24845553 17615350 30249741 29403074 22992520 30382153 18563158 23254933 20010809 19892987 25243066 22722832 14702039 12508121 15489334 21269460 22362889 22398555 17975172 16136131 21709235 21993624 21993625 26763976 27762356 28087693 23258410 23624526 17495919
EMBL
BABH01040265
KQ459585
KPI98182.1
KQ460627
KPJ13451.1
NWSH01002456
+ More
PCG68259.1 RSAL01000016 RVE53033.1 GBYB01014560 GBYB01014561 GBYB01014562 GBYB01014563 JAG84327.1 JAG84328.1 JAG84329.1 JAG84330.1 GEZM01039899 JAV81102.1 GL767538 EFZ13486.1 NNAY01002367 OXU21450.1 AAZX01000213 KQ982851 KYQ49830.1 KQ981567 KYN39802.1 KQ979074 KYN23050.1 ADTU01003356 KQ434886 KZC10113.1 KQ976692 KYM77686.1 KK107159 EZA56647.1 GL888336 EGI62489.1 GL446691 EFN87398.1 NEVH01007823 PNF35238.1 GL435242 EFN73624.1 KQ414648 KOC66156.1 KQ971338 EFA01806.1 KQ977622 KYN01343.1 GECU01037618 GECU01028402 JAS70088.1 JAS79304.1 GEZM01039897 JAV81106.1 KQ435740 KOX76845.1 DS235172 EEB12967.1 GECZ01024577 JAS45192.1 PNF35240.1 GEDC01013215 JAS24083.1 KZ288202 PBC33497.1 KK852567 KDR21227.1 KQ759874 OAD62374.1 DS469531 EDO45981.1 GEBQ01024646 JAT15331.1 QOIP01000001 RLU26717.1 HACG01032407 CEK79272.1 PYGN01000086 PSN54645.1 JH818965 EKC17980.1 RCHS01002309 RMX48136.1 GG666464 EEN68468.1 KB203854 ESO82669.1 JH431796 HACG01032403 CEK79268.1 GEZM01039900 GEZM01039898 JAV81100.1 ACTA01148253 ACTA01156252 HAAF01001938 CCP73764.1 GAMT01007352 GAMS01010919 GAMR01003050 GAMQ01002820 GAMP01002666 JAB04509.1 JAB12217.1 JAB30882.1 JAB39031.1 JAB50089.1 AJFE02037282 AJFE02037283 AK074553 AL356019 BC008647 BC030822 BC053538 CABD030091995 CABD030091996 CABD030091997 AANG04001761 AACZ04052400 GABC01010837 GABF01002296 GABD01010185 GABE01004007 NBAG03000456 JAA00501.1 JAA19849.1 JAA22915.1 JAA40732.1 PNI22209.1 AEFK01008730 AEFK01008731 AEFK01008732 ABGA01395405 NDHI03003485 PNJ36182.1 HACG01032409 CEK79274.1 KE162312 EPQ07737.1 AAPE02015125 KM486791 JH168595 AIZ09076.1 EHB04614.1 AQIB01128679 ADFV01127204 BC126001 CM004466 AAI26002.1 OCU01550.1 BC070853 AAH70853.1 AGTP01114621 AGTP01114622 AGTP01114623 AGTP01114624 KB102338 ELK35507.1 RQTK01000186 RUS84991.1 GFFW01001512 JAV43276.1 KB030984 ELK07090.1 KN124097 KFO22260.1 GABZ01006145 JAA47380.1 AEYP01102348 AEYP01102349 KB535354 EMP33620.1 AAKN02019211 AAKN02019212
PCG68259.1 RSAL01000016 RVE53033.1 GBYB01014560 GBYB01014561 GBYB01014562 GBYB01014563 JAG84327.1 JAG84328.1 JAG84329.1 JAG84330.1 GEZM01039899 JAV81102.1 GL767538 EFZ13486.1 NNAY01002367 OXU21450.1 AAZX01000213 KQ982851 KYQ49830.1 KQ981567 KYN39802.1 KQ979074 KYN23050.1 ADTU01003356 KQ434886 KZC10113.1 KQ976692 KYM77686.1 KK107159 EZA56647.1 GL888336 EGI62489.1 GL446691 EFN87398.1 NEVH01007823 PNF35238.1 GL435242 EFN73624.1 KQ414648 KOC66156.1 KQ971338 EFA01806.1 KQ977622 KYN01343.1 GECU01037618 GECU01028402 JAS70088.1 JAS79304.1 GEZM01039897 JAV81106.1 KQ435740 KOX76845.1 DS235172 EEB12967.1 GECZ01024577 JAS45192.1 PNF35240.1 GEDC01013215 JAS24083.1 KZ288202 PBC33497.1 KK852567 KDR21227.1 KQ759874 OAD62374.1 DS469531 EDO45981.1 GEBQ01024646 JAT15331.1 QOIP01000001 RLU26717.1 HACG01032407 CEK79272.1 PYGN01000086 PSN54645.1 JH818965 EKC17980.1 RCHS01002309 RMX48136.1 GG666464 EEN68468.1 KB203854 ESO82669.1 JH431796 HACG01032403 CEK79268.1 GEZM01039900 GEZM01039898 JAV81100.1 ACTA01148253 ACTA01156252 HAAF01001938 CCP73764.1 GAMT01007352 GAMS01010919 GAMR01003050 GAMQ01002820 GAMP01002666 JAB04509.1 JAB12217.1 JAB30882.1 JAB39031.1 JAB50089.1 AJFE02037282 AJFE02037283 AK074553 AL356019 BC008647 BC030822 BC053538 CABD030091995 CABD030091996 CABD030091997 AANG04001761 AACZ04052400 GABC01010837 GABF01002296 GABD01010185 GABE01004007 NBAG03000456 JAA00501.1 JAA19849.1 JAA22915.1 JAA40732.1 PNI22209.1 AEFK01008730 AEFK01008731 AEFK01008732 ABGA01395405 NDHI03003485 PNJ36182.1 HACG01032409 CEK79274.1 KE162312 EPQ07737.1 AAPE02015125 KM486791 JH168595 AIZ09076.1 EHB04614.1 AQIB01128679 ADFV01127204 BC126001 CM004466 AAI26002.1 OCU01550.1 BC070853 AAH70853.1 AGTP01114621 AGTP01114622 AGTP01114623 AGTP01114624 KB102338 ELK35507.1 RQTK01000186 RUS84991.1 GFFW01001512 JAV43276.1 KB030984 ELK07090.1 KN124097 KFO22260.1 GABZ01006145 JAA47380.1 AEYP01102348 AEYP01102349 KB535354 EMP33620.1 AAKN02019211 AAKN02019212
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000283053
UP000215335
+ More
UP000002358 UP000075809 UP000078541 UP000078492 UP000005205 UP000076502 UP000078540 UP000053097 UP000007755 UP000008237 UP000235965 UP000005203 UP000000311 UP000053825 UP000007266 UP000078542 UP000053105 UP000009046 UP000242457 UP000027135 UP000001593 UP000279307 UP000245037 UP000005408 UP000275408 UP000001554 UP000030746 UP000079169 UP000085678 UP000189704 UP000008912 UP000286642 UP000261680 UP000291022 UP000233220 UP000002281 UP000007646 UP000008225 UP000240080 UP000005640 UP000001519 UP000011712 UP000002277 UP000007648 UP000001595 UP000233040 UP000001074 UP000006813 UP000029965 UP000233020 UP000001073 UP000186698 UP000261681 UP000005215 UP000248480 UP000245340 UP000271974 UP000233140 UP000286641 UP000010552 UP000028990 UP000000715 UP000233060 UP000031443 UP000005447 UP000002280
UP000002358 UP000075809 UP000078541 UP000078492 UP000005205 UP000076502 UP000078540 UP000053097 UP000007755 UP000008237 UP000235965 UP000005203 UP000000311 UP000053825 UP000007266 UP000078542 UP000053105 UP000009046 UP000242457 UP000027135 UP000001593 UP000279307 UP000245037 UP000005408 UP000275408 UP000001554 UP000030746 UP000079169 UP000085678 UP000189704 UP000008912 UP000286642 UP000261680 UP000291022 UP000233220 UP000002281 UP000007646 UP000008225 UP000240080 UP000005640 UP000001519 UP000011712 UP000002277 UP000007648 UP000001595 UP000233040 UP000001074 UP000006813 UP000029965 UP000233020 UP000001073 UP000186698 UP000261681 UP000005215 UP000248480 UP000245340 UP000271974 UP000233140 UP000286641 UP000010552 UP000028990 UP000000715 UP000233060 UP000031443 UP000005447 UP000002280
Pfam
Interpro
IPR011990
TPR-like_helical_dom_sf
+ More
IPR032076 TTC5_OB
IPR019734 TPR_repeat
IPR038645 TTC5_OB_sf
IPR013026 TPR-contain_dom
IPR013105 TPR_2
IPR002870 Peptidase_M12B_N
IPR041645 ADAM_CR_2
IPR024079 MetalloPept_cat_dom_sf
IPR013273 ADAMTS/ADAMTS-like
IPR000884 TSP1_rpt
IPR036383 TSP1_rpt_sf
IPR010294 ADAM_spacer1
IPR001590 Peptidase_M12B
IPR032076 TTC5_OB
IPR019734 TPR_repeat
IPR038645 TTC5_OB_sf
IPR013026 TPR-contain_dom
IPR013105 TPR_2
IPR002870 Peptidase_M12B_N
IPR041645 ADAM_CR_2
IPR024079 MetalloPept_cat_dom_sf
IPR013273 ADAMTS/ADAMTS-like
IPR000884 TSP1_rpt
IPR036383 TSP1_rpt_sf
IPR010294 ADAM_spacer1
IPR001590 Peptidase_M12B
Gene 3D
ProteinModelPortal
H9JJ39
A0A194PXX8
A0A194RC17
A0A2A4J8R8
A0A3S2P5N7
A0A0C9RYX1
+ More
A0A1Y1M5P0 E9J0S8 A0A232ESW6 K7J6R9 A0A151WPS4 A0A151JXG6 A0A195ECV5 A0A158NXR6 A0A154PE41 A0A195AZQ6 A0A026WKS7 F4WTH1 E2BAA0 A0A2J7R321 A0A087ZRS6 E1ZYT4 A0A0L7R5W3 D1ZZR8 A0A195CKV9 A0A1B6HXA3 A0A1Y1MB57 A0A0M9A450 E0VHW1 A0A1B6F4K9 A0A2J7R335 A0A1B6DEG0 A0A2A3EP51 A0A067RBV7 A0A310SWV8 A7RRB4 A0A1B6KVA8 A0A3L8E224 A0A0B7AF55 A0A2P8ZDS4 K1PNB1 A0A3M6U3G2 C3XU94 V4B2K3 T1J2A1 A0A3Q0JKL5 A0A1S3JSL4 A0A1S3JT74 A0A0B7AEN7 A0A1Y1M944 A0A3Q0E0Q4 G1LG62 A0A1W2W544 A0A3Q7VBL8 A0A3Q0K5Y5 A0A384BH51 A0A452RHI0 A0A2K6U289 F6Q337 U6CTX2 G3SQJ0 U3C0A8 F7D0C6 A0A2R9BP76 Q8N0Z6 G3QDN9 M3W130 A0A2I3RW55 G3WGM9 H2NKH1 A0A2K5QJJ6 A0A0B7AE47 S7PHZ4 G1PS04 G5B5Q3 A0A0D9RW71 A0A2K5F1G4 A0A2I3H703 A0JMT7 Q6NRA9 A0A383ZZM7 I3MEN8 A0A2Y9EAG2 A0A2U3WUI0 L5MB24 A0A3S1BJB2 A0A2K5YA58 A0A3Q7RI40 A0A250YIA0 L5K6D8 A0A091CWR2 K9IJW8 M3Z354 A0A2K5M0H4 M7BDF6 H0VDM7 F6WBF3
A0A1Y1M5P0 E9J0S8 A0A232ESW6 K7J6R9 A0A151WPS4 A0A151JXG6 A0A195ECV5 A0A158NXR6 A0A154PE41 A0A195AZQ6 A0A026WKS7 F4WTH1 E2BAA0 A0A2J7R321 A0A087ZRS6 E1ZYT4 A0A0L7R5W3 D1ZZR8 A0A195CKV9 A0A1B6HXA3 A0A1Y1MB57 A0A0M9A450 E0VHW1 A0A1B6F4K9 A0A2J7R335 A0A1B6DEG0 A0A2A3EP51 A0A067RBV7 A0A310SWV8 A7RRB4 A0A1B6KVA8 A0A3L8E224 A0A0B7AF55 A0A2P8ZDS4 K1PNB1 A0A3M6U3G2 C3XU94 V4B2K3 T1J2A1 A0A3Q0JKL5 A0A1S3JSL4 A0A1S3JT74 A0A0B7AEN7 A0A1Y1M944 A0A3Q0E0Q4 G1LG62 A0A1W2W544 A0A3Q7VBL8 A0A3Q0K5Y5 A0A384BH51 A0A452RHI0 A0A2K6U289 F6Q337 U6CTX2 G3SQJ0 U3C0A8 F7D0C6 A0A2R9BP76 Q8N0Z6 G3QDN9 M3W130 A0A2I3RW55 G3WGM9 H2NKH1 A0A2K5QJJ6 A0A0B7AE47 S7PHZ4 G1PS04 G5B5Q3 A0A0D9RW71 A0A2K5F1G4 A0A2I3H703 A0JMT7 Q6NRA9 A0A383ZZM7 I3MEN8 A0A2Y9EAG2 A0A2U3WUI0 L5MB24 A0A3S1BJB2 A0A2K5YA58 A0A3Q7RI40 A0A250YIA0 L5K6D8 A0A091CWR2 K9IJW8 M3Z354 A0A2K5M0H4 M7BDF6 H0VDM7 F6WBF3
PDB
4ABN
E-value=4.03503e-69,
Score=665
Ontologies
GO
Topology
Subcellular location
Nucleus
Phosphorylation at Ser-203 results in nuclear localization, while unphosphorylated protein localizes to the cytoplasm. With evidence from 2 publications.
Cytoplasm Phosphorylation at Ser-203 results in nuclear localization, while unphosphorylated protein localizes to the cytoplasm. With evidence from 2 publications.
Cytoplasm Phosphorylation at Ser-203 results in nuclear localization, while unphosphorylated protein localizes to the cytoplasm. With evidence from 2 publications.
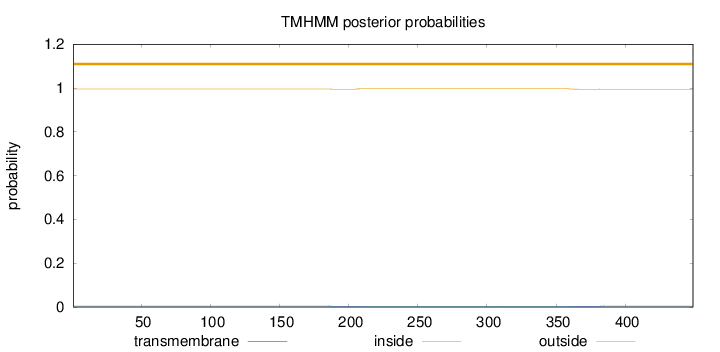
Length:
449
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10117
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00491
outside
1 - 449
Population Genetic Test Statistics
Pi
288.889787
Theta
196.956525
Tajima's D
1.805276
CLR
0.010904
CSRT
0.850007499625019
Interpretation
Uncertain
