Gene
KWMTBOMO08287
Pre Gene Modal
BGIBMGA009532
Annotation
PREDICTED:_torso-like_protein_isoform_X1_[Bombyx_mori]
Full name
Torso-like protein
Location in the cell
PlasmaMembrane Reliability : 2.387
Sequence
CDS
ATGGTGCGGACCCTCACAGTTCTGCTGCTGGTGGTAGCTCTGTGCCGCGCTGCCACCCATGACTCAGAGCTTGGCGTCGGGAAGGCCATCAACATATTTATGAGATATGGCTACCTCAGCATATGCATGCGAGTTGTTCCCCGGAACGACACAGAGAACTGGGTTTTCCGGGAGCCGACAGTCAGCGTTTTCCGGGACGTGGAGCAATACAGCGTCGCGCCCAAACCTCGCCAGCCGAAGACACTGTTCGACGGAGACTTCCACATGGAGTTCTGCGATAACCTGAAGCAACTGCTGCAAGCGTATTTCAGGGATTTCACTTTTGAAAGGCTGGAGCGTCCATGGCGGGCGTTCACGGCTGGCTGGCCCACAGACATAATGGCCAGAAACTTGGGGATCAACTCCTCGTTCATCAATGGCGACCACTGCTACGTGCTCGTCAGAGTTTCCAGGTTCCGCGAAACGGGAAAACTAAAGGACTTGCCACAGAACATAGGAGTTGAGGATGTGGTGTTTGACGCCATCCAAGATTCAATCATTGGGGATACGGCATCCATTGCCGATTTCATACGCAAATACGGATCGCACTACATTGCATCCTATATTACTGGTAACTCTCTGTACCAGGTGTTTGTCTTCTCCCGGGGCGTGTACTCCAGGGTCAAAGACCGCGTCAAGTCTAACGGGGTCGGTGACCTATCTGTCACTGAGCTAAACAATTTCTTCTCGCCGTTCTTCGCTGAACACGTCGGAGTAGTCAAGGTCGCCAGCGGTAACAAAACAGTAGAGAGCTGGGCGACGAAACGTCTAAGGATGCATTACTATATCTTCTCATACCCGAGTCTCCTGAAACTCCACGGGGAACCGGCGTTACTACGCAACCTAGACACGTTATTAGGAAATGAAGCTCTATTGCAATTGGAATTAAAGACATTATCGCCCGCATTCAAAGACGCGAACAAAAGGAAGTGGTTCGAAGAGGTCATAGACAATTATTTAAAACTTTGGGAGAGTAATATGTGA
Protein
MVRTLTVLLLVVALCRAATHDSELGVGKAINIFMRYGYLSICMRVVPRNDTENWVFREPTVSVFRDVEQYSVAPKPRQPKTLFDGDFHMEFCDNLKQLLQAYFRDFTFERLERPWRAFTAGWPTDIMARNLGINSSFINGDHCYVLVRVSRFRETGKLKDLPQNIGVEDVVFDAIQDSIIGDTASIADFIRKYGSHYIASYITGNSLYQVFVFSRGVYSRVKDRVKSNGVGDLSVTELNNFFSPFFAEHVGVVKVASGNKTVESWATKRLRMHYYIFSYPSLLKLHGEPALLRNLDTLLGNEALLQLELKTLSPAFKDANKRKWFEEVIDNYLKLWESNM
Summary
Description
Probable ligand that binds to the torso receptor. Implicated in a receptor tyrosine kinase signaling pathway that specifies terminal cell fate.
Keywords
Complete proteome
Developmental protein
Glycoprotein
Reference proteome
Secreted
Signal
Feature
chain Torso-like protein
Uniprot
H9JJ31
A0A437BRT7
A0A2A4J9A2
A0A194Q444
A0A194R7F6
A0A2H1VDX1
+ More
A0A2H1VYN4 A0A212F771 A0A0L7L232 A0A087ZRM0 A0A1Y1M5U3 A0A151JXF1 A0A2A3EQJ3 E1ZYT3 A0A310SMA8 A0A195B067 A0A158NXR4 F4WTH2 A0A195ECW4 A0A195CMN1 A0A151WPU8 E2BA99 A0A026WLY0 K7J6S0 A0A3L8E1V4 A0A154PF96 D1ZZR7 A0A232ESY5 A0A0J7KT47 A0A2P8ZDR9 A0A0L7R5J4 A0A1L8DBU3 A0A1L8DNX8 A0A0M9A5G1 A0A1B0CAV8 A0A1L8DCJ7 A0A067RDY9 J3JYR3 A0A336KKX2 A0A1W4XTA4 A0A1W4XTF2 A0A2J7R311 A0A2S2PY11 N6TJ77 A0A182GFC4 Q16SW4 A0A1S4FQD7 A0A2S2P0R3 A0A2M3ZD38 A0A2H8TCZ6 A0A1J1HNJ1 W5JQF1 A0A182Q629 A0A182NG74 A0A182FPY3 A0A084W9E7 A0A182J0Z6 J9K914 A0A1B6EJS8 N6UTX8 A0A1I8PVB2 A0A182KG80 E0VBB4 A0A1B6KRP8 A0A182HS14 A0A182TZW2 A0A1A9UNM9 A0A182LQC4 A0A182USV6 A0A182XLW6 Q7QKE1 A0A182PEG9 A0A1A9ZEC0 A0A182R7M6 A0A1B0FLR4 A0A1A9YQU7 A0A1B0BGX2 B4MC56 A0A1A9X2M4 A0A3B0JL52 A0A182W2K2 Q29AR5 A0A182XVC4 B4G608 A0A1W4W3E5 A0A182M1F3 B4NHN7 A0A0K8UBG9 A0A034W306 B3LVH4 A0A0M4EE08 A0A1I8N3D6 B4K702 B4HM45 B4QZ92 A0A0A9YTX1 A4V379 P40689 B4JGQ2
A0A2H1VYN4 A0A212F771 A0A0L7L232 A0A087ZRM0 A0A1Y1M5U3 A0A151JXF1 A0A2A3EQJ3 E1ZYT3 A0A310SMA8 A0A195B067 A0A158NXR4 F4WTH2 A0A195ECW4 A0A195CMN1 A0A151WPU8 E2BA99 A0A026WLY0 K7J6S0 A0A3L8E1V4 A0A154PF96 D1ZZR7 A0A232ESY5 A0A0J7KT47 A0A2P8ZDR9 A0A0L7R5J4 A0A1L8DBU3 A0A1L8DNX8 A0A0M9A5G1 A0A1B0CAV8 A0A1L8DCJ7 A0A067RDY9 J3JYR3 A0A336KKX2 A0A1W4XTA4 A0A1W4XTF2 A0A2J7R311 A0A2S2PY11 N6TJ77 A0A182GFC4 Q16SW4 A0A1S4FQD7 A0A2S2P0R3 A0A2M3ZD38 A0A2H8TCZ6 A0A1J1HNJ1 W5JQF1 A0A182Q629 A0A182NG74 A0A182FPY3 A0A084W9E7 A0A182J0Z6 J9K914 A0A1B6EJS8 N6UTX8 A0A1I8PVB2 A0A182KG80 E0VBB4 A0A1B6KRP8 A0A182HS14 A0A182TZW2 A0A1A9UNM9 A0A182LQC4 A0A182USV6 A0A182XLW6 Q7QKE1 A0A182PEG9 A0A1A9ZEC0 A0A182R7M6 A0A1B0FLR4 A0A1A9YQU7 A0A1B0BGX2 B4MC56 A0A1A9X2M4 A0A3B0JL52 A0A182W2K2 Q29AR5 A0A182XVC4 B4G608 A0A1W4W3E5 A0A182M1F3 B4NHN7 A0A0K8UBG9 A0A034W306 B3LVH4 A0A0M4EE08 A0A1I8N3D6 B4K702 B4HM45 B4QZ92 A0A0A9YTX1 A4V379 P40689 B4JGQ2
Pubmed
19121390
26354079
22118469
26227816
28004739
20798317
+ More
21347285 21719571 24508170 20075255 30249741 18362917 19820115 28648823 29403074 24845553 22516182 23537049 26483478 17510324 20920257 23761445 24438588 20566863 20966253 12364791 14747013 17210077 17994087 15632085 23185243 25244985 25348373 25315136 18057021 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8107870 8276237 12537569 12814553 17893096
21347285 21719571 24508170 20075255 30249741 18362917 19820115 28648823 29403074 24845553 22516182 23537049 26483478 17510324 20920257 23761445 24438588 20566863 20966253 12364791 14747013 17210077 17994087 15632085 23185243 25244985 25348373 25315136 18057021 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8107870 8276237 12537569 12814553 17893096
EMBL
BABH01023289
RSAL01000016
RVE53031.1
NWSH01002456
PCG68258.1
KQ459585
+ More
KPI98180.1 KQ460627 KPJ13449.1 ODYU01002028 SOQ39025.1 ODYU01005267 SOQ45949.1 AGBW02009914 OWR49580.1 JTDY01003572 KOB69324.1 GEZM01039900 GEZM01039899 GEZM01039898 GEZM01039897 JAV81103.1 KQ981567 KYN39803.1 KZ288202 PBC33496.1 GL435242 EFN73623.1 KQ759874 OAD62375.1 KQ976692 KYM77687.1 ADTU01003356 GL888336 EGI62490.1 KQ979074 KYN23048.1 KQ977622 KYN01344.1 KQ982851 KYQ49831.1 GL446691 EFN87397.1 KK107159 EZA56646.1 AAZX01000213 QOIP01000001 RLU26710.1 KQ434886 KZC10114.1 KQ971338 EFA02884.1 NNAY01002367 OXU21452.1 LBMM01003459 KMQ93508.1 PYGN01000086 PSN54644.1 KQ414648 KOC66155.1 GFDF01010148 JAV03936.1 GFDF01005922 JAV08162.1 KQ435740 KOX76846.1 AJWK01004316 GFDF01009896 JAV04188.1 KK852567 KDR21228.1 BT128393 KB632308 AEE63351.1 ERL92003.1 UFQS01000555 UFQT01000555 SSX04891.1 SSX25254.1 NEVH01007823 PNF35228.1 GGMS01001193 MBY70396.1 APGK01023865 APGK01023866 APGK01023867 KB740522 ENN80499.1 JXUM01000913 JXUM01016155 JXUM01016156 JXUM01016157 KQ560450 KQ560113 KXJ82365.1 KXJ84426.1 CH477664 EAT37555.1 GGMR01010418 MBY23037.1 GGFM01005703 MBW26454.1 GFXV01000155 GFXV01006756 MBW11960.1 MBW18561.1 CVRI01000010 CRK89098.1 ADMH02000471 ETN66351.1 AXCN02001631 ATLV01021724 KE525322 KFB46841.1 ABLF02031261 ABLF02031262 GECZ01031627 JAS38142.1 APGK01015109 APGK01015110 KB739485 ENN82247.1 DS235023 EEB10670.1 GEBQ01026063 JAT13914.1 APCN01000232 AAAB01008799 EAA03773.4 CCAG010015430 JXJN01014062 CH940656 EDW58677.1 OUUW01000007 SPP82955.1 CM000070 EAL27284.2 KRS99906.1 KRS99907.1 KRS99908.1 CH479179 EDW23775.1 AXCM01000557 CH964272 EDW84647.1 GDHF01028297 JAI24017.1 GAKP01010799 JAC48153.1 CH902617 EDV42544.1 CP012526 ALC46154.1 CH933806 EDW15289.1 KRG01497.1 KRG01498.1 KRG01499.1 CH480815 EDW43093.1 CM000364 EDX13828.1 GBHO01008538 GBRD01016118 GDHC01016940 JAG35066.1 JAG49708.1 JAQ01689.1 AE014297 AAN13883.1 AGB96190.1 Z30342 X75614 AY071336 CH916369 EDV92656.1
KPI98180.1 KQ460627 KPJ13449.1 ODYU01002028 SOQ39025.1 ODYU01005267 SOQ45949.1 AGBW02009914 OWR49580.1 JTDY01003572 KOB69324.1 GEZM01039900 GEZM01039899 GEZM01039898 GEZM01039897 JAV81103.1 KQ981567 KYN39803.1 KZ288202 PBC33496.1 GL435242 EFN73623.1 KQ759874 OAD62375.1 KQ976692 KYM77687.1 ADTU01003356 GL888336 EGI62490.1 KQ979074 KYN23048.1 KQ977622 KYN01344.1 KQ982851 KYQ49831.1 GL446691 EFN87397.1 KK107159 EZA56646.1 AAZX01000213 QOIP01000001 RLU26710.1 KQ434886 KZC10114.1 KQ971338 EFA02884.1 NNAY01002367 OXU21452.1 LBMM01003459 KMQ93508.1 PYGN01000086 PSN54644.1 KQ414648 KOC66155.1 GFDF01010148 JAV03936.1 GFDF01005922 JAV08162.1 KQ435740 KOX76846.1 AJWK01004316 GFDF01009896 JAV04188.1 KK852567 KDR21228.1 BT128393 KB632308 AEE63351.1 ERL92003.1 UFQS01000555 UFQT01000555 SSX04891.1 SSX25254.1 NEVH01007823 PNF35228.1 GGMS01001193 MBY70396.1 APGK01023865 APGK01023866 APGK01023867 KB740522 ENN80499.1 JXUM01000913 JXUM01016155 JXUM01016156 JXUM01016157 KQ560450 KQ560113 KXJ82365.1 KXJ84426.1 CH477664 EAT37555.1 GGMR01010418 MBY23037.1 GGFM01005703 MBW26454.1 GFXV01000155 GFXV01006756 MBW11960.1 MBW18561.1 CVRI01000010 CRK89098.1 ADMH02000471 ETN66351.1 AXCN02001631 ATLV01021724 KE525322 KFB46841.1 ABLF02031261 ABLF02031262 GECZ01031627 JAS38142.1 APGK01015109 APGK01015110 KB739485 ENN82247.1 DS235023 EEB10670.1 GEBQ01026063 JAT13914.1 APCN01000232 AAAB01008799 EAA03773.4 CCAG010015430 JXJN01014062 CH940656 EDW58677.1 OUUW01000007 SPP82955.1 CM000070 EAL27284.2 KRS99906.1 KRS99907.1 KRS99908.1 CH479179 EDW23775.1 AXCM01000557 CH964272 EDW84647.1 GDHF01028297 JAI24017.1 GAKP01010799 JAC48153.1 CH902617 EDV42544.1 CP012526 ALC46154.1 CH933806 EDW15289.1 KRG01497.1 KRG01498.1 KRG01499.1 CH480815 EDW43093.1 CM000364 EDX13828.1 GBHO01008538 GBRD01016118 GDHC01016940 JAG35066.1 JAG49708.1 JAQ01689.1 AE014297 AAN13883.1 AGB96190.1 Z30342 X75614 AY071336 CH916369 EDV92656.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000005203 UP000078541 UP000242457 UP000000311 UP000078540 UP000005205 UP000007755 UP000078492 UP000078542 UP000075809 UP000008237 UP000053097 UP000002358 UP000279307 UP000076502 UP000007266 UP000215335 UP000036403 UP000245037 UP000053825 UP000053105 UP000092461 UP000027135 UP000030742 UP000192223 UP000235965 UP000019118 UP000069940 UP000249989 UP000008820 UP000183832 UP000000673 UP000075886 UP000075884 UP000069272 UP000030765 UP000075880 UP000007819 UP000095300 UP000075881 UP000009046 UP000075840 UP000075902 UP000078200 UP000075882 UP000075903 UP000076407 UP000007062 UP000075885 UP000092445 UP000075900 UP000092444 UP000092443 UP000092460 UP000008792 UP000091820 UP000268350 UP000075920 UP000001819 UP000076408 UP000008744 UP000192221 UP000075883 UP000007798 UP000007801 UP000092553 UP000095301 UP000009192 UP000001292 UP000000304 UP000000803 UP000001070
UP000037510 UP000005203 UP000078541 UP000242457 UP000000311 UP000078540 UP000005205 UP000007755 UP000078492 UP000078542 UP000075809 UP000008237 UP000053097 UP000002358 UP000279307 UP000076502 UP000007266 UP000215335 UP000036403 UP000245037 UP000053825 UP000053105 UP000092461 UP000027135 UP000030742 UP000192223 UP000235965 UP000019118 UP000069940 UP000249989 UP000008820 UP000183832 UP000000673 UP000075886 UP000075884 UP000069272 UP000030765 UP000075880 UP000007819 UP000095300 UP000075881 UP000009046 UP000075840 UP000075902 UP000078200 UP000075882 UP000075903 UP000076407 UP000007062 UP000075885 UP000092445 UP000075900 UP000092444 UP000092443 UP000092460 UP000008792 UP000091820 UP000268350 UP000075920 UP000001819 UP000076408 UP000008744 UP000192221 UP000075883 UP000007798 UP000007801 UP000092553 UP000095301 UP000009192 UP000001292 UP000000304 UP000000803 UP000001070
Pfam
PF01823 MACPF
Interpro
IPR020864
MACPF
ProteinModelPortal
H9JJ31
A0A437BRT7
A0A2A4J9A2
A0A194Q444
A0A194R7F6
A0A2H1VDX1
+ More
A0A2H1VYN4 A0A212F771 A0A0L7L232 A0A087ZRM0 A0A1Y1M5U3 A0A151JXF1 A0A2A3EQJ3 E1ZYT3 A0A310SMA8 A0A195B067 A0A158NXR4 F4WTH2 A0A195ECW4 A0A195CMN1 A0A151WPU8 E2BA99 A0A026WLY0 K7J6S0 A0A3L8E1V4 A0A154PF96 D1ZZR7 A0A232ESY5 A0A0J7KT47 A0A2P8ZDR9 A0A0L7R5J4 A0A1L8DBU3 A0A1L8DNX8 A0A0M9A5G1 A0A1B0CAV8 A0A1L8DCJ7 A0A067RDY9 J3JYR3 A0A336KKX2 A0A1W4XTA4 A0A1W4XTF2 A0A2J7R311 A0A2S2PY11 N6TJ77 A0A182GFC4 Q16SW4 A0A1S4FQD7 A0A2S2P0R3 A0A2M3ZD38 A0A2H8TCZ6 A0A1J1HNJ1 W5JQF1 A0A182Q629 A0A182NG74 A0A182FPY3 A0A084W9E7 A0A182J0Z6 J9K914 A0A1B6EJS8 N6UTX8 A0A1I8PVB2 A0A182KG80 E0VBB4 A0A1B6KRP8 A0A182HS14 A0A182TZW2 A0A1A9UNM9 A0A182LQC4 A0A182USV6 A0A182XLW6 Q7QKE1 A0A182PEG9 A0A1A9ZEC0 A0A182R7M6 A0A1B0FLR4 A0A1A9YQU7 A0A1B0BGX2 B4MC56 A0A1A9X2M4 A0A3B0JL52 A0A182W2K2 Q29AR5 A0A182XVC4 B4G608 A0A1W4W3E5 A0A182M1F3 B4NHN7 A0A0K8UBG9 A0A034W306 B3LVH4 A0A0M4EE08 A0A1I8N3D6 B4K702 B4HM45 B4QZ92 A0A0A9YTX1 A4V379 P40689 B4JGQ2
A0A2H1VYN4 A0A212F771 A0A0L7L232 A0A087ZRM0 A0A1Y1M5U3 A0A151JXF1 A0A2A3EQJ3 E1ZYT3 A0A310SMA8 A0A195B067 A0A158NXR4 F4WTH2 A0A195ECW4 A0A195CMN1 A0A151WPU8 E2BA99 A0A026WLY0 K7J6S0 A0A3L8E1V4 A0A154PF96 D1ZZR7 A0A232ESY5 A0A0J7KT47 A0A2P8ZDR9 A0A0L7R5J4 A0A1L8DBU3 A0A1L8DNX8 A0A0M9A5G1 A0A1B0CAV8 A0A1L8DCJ7 A0A067RDY9 J3JYR3 A0A336KKX2 A0A1W4XTA4 A0A1W4XTF2 A0A2J7R311 A0A2S2PY11 N6TJ77 A0A182GFC4 Q16SW4 A0A1S4FQD7 A0A2S2P0R3 A0A2M3ZD38 A0A2H8TCZ6 A0A1J1HNJ1 W5JQF1 A0A182Q629 A0A182NG74 A0A182FPY3 A0A084W9E7 A0A182J0Z6 J9K914 A0A1B6EJS8 N6UTX8 A0A1I8PVB2 A0A182KG80 E0VBB4 A0A1B6KRP8 A0A182HS14 A0A182TZW2 A0A1A9UNM9 A0A182LQC4 A0A182USV6 A0A182XLW6 Q7QKE1 A0A182PEG9 A0A1A9ZEC0 A0A182R7M6 A0A1B0FLR4 A0A1A9YQU7 A0A1B0BGX2 B4MC56 A0A1A9X2M4 A0A3B0JL52 A0A182W2K2 Q29AR5 A0A182XVC4 B4G608 A0A1W4W3E5 A0A182M1F3 B4NHN7 A0A0K8UBG9 A0A034W306 B3LVH4 A0A0M4EE08 A0A1I8N3D6 B4K702 B4HM45 B4QZ92 A0A0A9YTX1 A4V379 P40689 B4JGQ2
Ontologies
GO
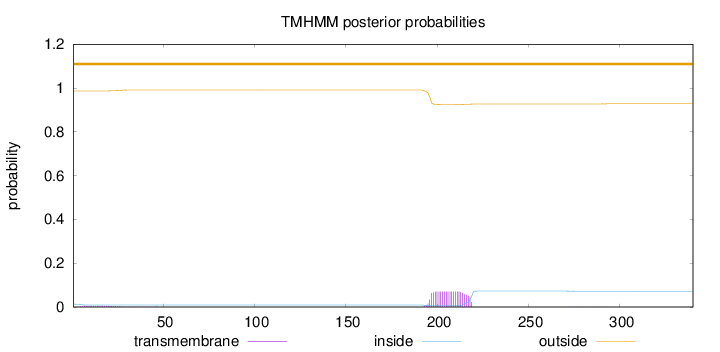
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 17,
Likelihood: 0.996617
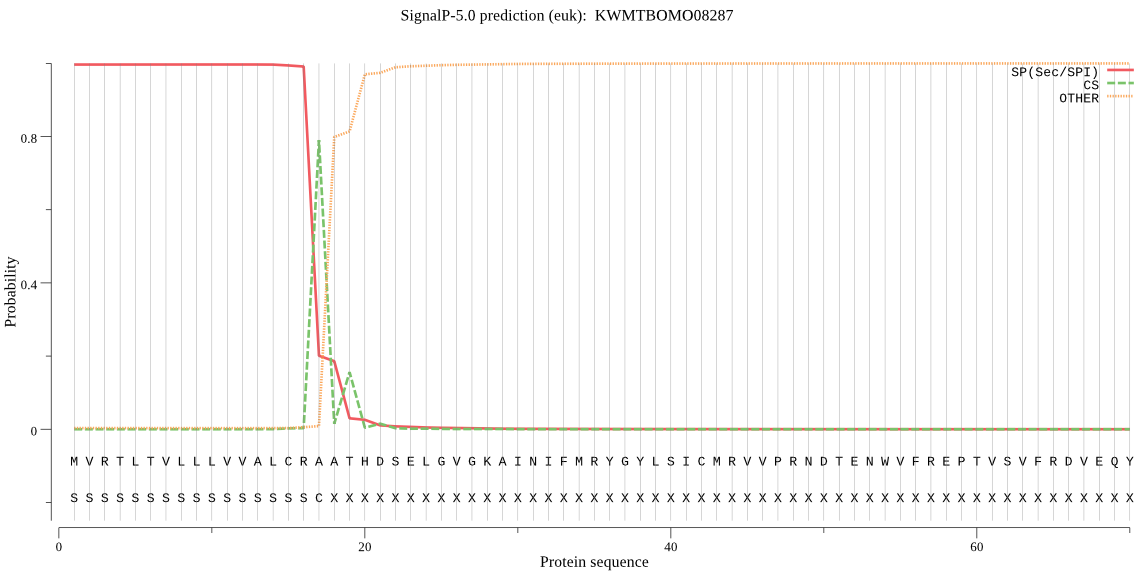
Length:
340
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.66377
Exp number, first 60 AAs:
0.10833
Total prob of N-in:
0.01347
outside
1 - 340
Population Genetic Test Statistics
Pi
245.896669
Theta
206.540439
Tajima's D
1.791827
CLR
0.84092
CSRT
0.846357682115894
Interpretation
Uncertain
