Gene
KWMTBOMO08283
Pre Gene Modal
BGIBMGA009432
Annotation
PREDICTED:_paired_box_protein_Pax-6?_partial_[Amyelois_transitella]
Full name
Paired box protein Pax-6
Alternative Name
Oculorhombin
Aniridia type II protein
Aniridia type II protein
Transcription factor
Location in the cell
Nuclear Reliability : 4.736
Sequence
CDS
ATGCACAGCGCGGCTATGGGTGGTGGAGGCCTCTTCGGATGCTCGTCGGCTGGTCACAGCGGCATCAATCAGCTGGGCGGGGTCTACGTCAACGGACGACCGCTGCCGGACTCGACTCGGCAAAAGATCGTGGAACTAGCCCACTCCGGAGCTAGACCCTGCGACATCAGCCGCATCCTGCAGGTCTCCAACGGCTGTGTCTCCAAAATACTAGGCAGCTTCACACGCAGATATTACGAGACGGGGTCGATAAAGCCACGCGCGATTGGTGGGTCCAAGCCCCGGGTGGCGACCACTCCAGTTGTCCAGAAGATAGCCGACTACAAGCGGGAGTGCCCCTCGATCTTCGCCTGGGAGATCAGGGACCGCCTGCTCAGCGAAAACGTGTGCAATAACGACAACATACCCAGTGTGTCATCAATAAACCGAGTACTACGGAACCTGGCCTCGCAGAAGGAGCAAGCGGCATCAGCGCAGAACGATAGCGTGTATGAGAAACTGAGGATGTTCAATGGTCAAGCGGCGACTGGATGGTGGTATCCAGGGCTACCCACCGCGCCGACCGCACCGGCGATACCAGCCCCACTGCCCCCTCAACTCAGGACCACAACAGAAGAGCAGAAACGAGATACAATTCAGTCGGAAGCGGGTTCAGACGGGAACAGTGAACATGCGTCATCAGGCGACGAGGACTCGCAGATGAGACTCCGATTGAAGAGGAAACTCCAGAGGAACAGGACCAGTTTCACAAATGACCAAATAGATAGTCTCGAGAAGGAATTCGAGAGGACCCACTACCCGGACGTGTTCGCCCGGGAGAGACTCGCTGAAAAGATTGGATTGCCTGAGGCACGTATCCAGGTATGGTTCTCAAATCGTCGTGCAAAATGGCGTCGCGAAGAGAAACTCCGGAGCCAGCGTCGCGACGCGCCGGCCGCGTCTCCTCCCGCACCCCCCGCGCCGCGACTCCCCCTCAATGGAGGCTTCAATTCCATGTACAGCCCCATACCGCAACCCATTGCCACAATGGCTGATTCTTACAGCTCGATGTCTGGTGGGCTCTCCTCATCGTGTCTCCAGCAGCGAGACGGCGGCTACCCCTACATGTTCGGGGACGTCCTCGCCAGCGGAGGGTACTCCAGAGCCCCGGCAGCGCATCAGCAGCACCCGGCTTACTCACAGCCACAGCCCGCTGGCAGTACGGGTGTGATATCGGCGGGCGTGAGCGTACCCGTGCAGATACCGTCGCAGGGGCCCGACCTCGCGTCCAACTACTGGGGCCGGCTCCAGTGA
Protein
MHSAAMGGGGLFGCSSAGHSGINQLGGVYVNGRPLPDSTRQKIVELAHSGARPCDISRILQVSNGCVSKILGSFTRRYYETGSIKPRAIGGSKPRVATTPVVQKIADYKRECPSIFAWEIRDRLLSENVCNNDNIPSVSSINRVLRNLASQKEQAASAQNDSVYEKLRMFNGQAATGWWYPGLPTAPTAPAIPAPLPPQLRTTTEEQKRDTIQSEAGSDGNSEHASSGDEDSQMRLRLKRKLQRNRTSFTNDQIDSLEKEFERTHYPDVFARERLAEKIGLPEARIQVWFSNRRAKWRREEKLRSQRRDAPAASPPAPPAPRLPLNGGFNSMYSPIPQPIATMADSYSSMSGGLSSSCLQQRDGGYPYMFGDVLASGGYSRAPAAHQQHPAYSQPQPAGSTGVISAGVSVPVQIPSQGPDLASNYWGRLQ
Summary
Description
Transcription factor with important functions in the development of the eye, nose, central nervous system and pancreas. Required for the differentiation of pancreatic islet alpha cells. Competes with PAX4 in binding to a common element in the glucagon, insulin and somatostatin promoters. Regulates specification of the ventral neuron subtypes by establishing the correct progenitor domains (By similarity).
Transcription factor with important functions in the development of the eye, nose, central nervous system and pancreas. Required for the differentiation of pancreatic islet alpha cells (By similarity). Competes with PAX4 in binding to a common element in the glucagon, insulin and somatostatin promoters. Regulates specification of the ventral neuron subtypes by establishing the correct progenitor domains (By similarity). Isoform 5a appears to function as a molecular switch that specifies target genes.
Transcription factor with important functions in the development of the eye, nose, central nervous system and pancreas. Required for the differentiation of pancreatic islet alpha cells. Competes with PAX4 in binding to a common element in the glucagon, insulin and somatostatin promoters (By similarity). Regulates specification of the ventral neuron subtypes by establishing the correct progenitor domains.
Transcription factor with important functions in the development of the eye, nose, central nervous system and pancreas. Required for the differentiation of pancreatic islet alpha cells (By similarity). Competes with PAX4 in binding to a common element in the glucagon, insulin and somatostatin promoters. Regulates specification of the ventral neuron subtypes by establishing the correct progenitor domains (By similarity). Isoform 5a appears to function as a molecular switch that specifies target genes.
Transcription factor with important functions in the development of the eye, nose, central nervous system and pancreas. Required for the differentiation of pancreatic islet alpha cells. Competes with PAX4 in binding to a common element in the glucagon, insulin and somatostatin promoters (By similarity). Regulates specification of the ventral neuron subtypes by establishing the correct progenitor domains.
Subunit
Interacts with MAF and MAFB. Interacts with TRIM11; this interaction leads to ubiquitination and proteasomal degradation, as well as inhibition of transactivation, possibly in part by preventing PAX6 binding to consensus DNA sequences.
Similarity
Belongs to the paired homeobox family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Differentiation
Direct protein sequencing
Disease mutation
DNA-binding
Homeobox
Nucleus
Paired box
Reference proteome
Transcription
Transcription regulation
Ubl conjugation
3D-structure
Peters anomaly
Repressor
Feature
chain Paired box protein Pax-6
splice variant In isoform 3.
sequence variant In AN1.
splice variant In isoform 3.
sequence variant In AN1.
Uniprot
A0A194Q439
A0A194RC13
A0A3S2LG24
A0A212FLI3
D1ZZR4
A0A139WJ10
+ More
A0A2J7PYX8 A0A1B6D7L7 A0A1B6HV64 A0A336L552 A0A336LKB8 E0VCT1 A0A1J1HK27 E2BA95 A0A146LD26 A0A1D2N7X8 K7J6S2 A0A195CKV0 A0A067RSK7 A0A226E7T7 T1JIT2 A0A0N8EJN0 W8BQD7 W8BW79 A0A0F7TDH4 A0A3B0KXY6 B5DRZ9 B4HC93 B4JZP2 B4NHL0 B4MEY7 B4L7D3 A0A0L0CCV2 B4IKQ3 Q9V490 Q9XZC2 A0A0J9S132 A0A1W4VHQ6 B3P9Q4 A8VFE9 A0A0R3P0G3 A0A3B0K3Y7 B3N1J5 A0A286XBI8 Q38L56 G1T0K0 A0A287D9N3 A0A2K6CL21 A0A2K6L4C8 G1P774 A0A2I2ZFV9 A0A287AAF3 A0A2K5Y8D0 A0A2R9CK82 A0A2K5MMA0 H0XKU3 A0A2K5JGR6 H9FV32 A0A2K6S284 A0A2K6GSJ3 A0A3Q7XWK9 A0A2Y9D899 A0A2J8WG99 A0A2U3WCG5 I7G9J6 A0A2J8P422 F6S4R0 Q66SS1 P63015 P26367 A0A0Q9WKW4 B3KQG1 A0A1S3FEX4 P63016 G3TZY7 A0A2K5QU00 A0A3Q7U5D5 W5Q3U9 Q1LZF1 A0A341C1H6 A0A2Y9F604 A0A340XHU6 A0A384A1M6 A0A3Q0D361 H0WX77 A0A1U7S5R9 A0A0Q9XFH6 K7G268 A0A1V4JP70 A4GG27 G1LK37 D3DQZ8 A0A401SVE9 G1KAP0 A0A401P0Q4 A0A2K5Y8E0 L0MPR7 Q9I877 A0A0J9USS3
A0A2J7PYX8 A0A1B6D7L7 A0A1B6HV64 A0A336L552 A0A336LKB8 E0VCT1 A0A1J1HK27 E2BA95 A0A146LD26 A0A1D2N7X8 K7J6S2 A0A195CKV0 A0A067RSK7 A0A226E7T7 T1JIT2 A0A0N8EJN0 W8BQD7 W8BW79 A0A0F7TDH4 A0A3B0KXY6 B5DRZ9 B4HC93 B4JZP2 B4NHL0 B4MEY7 B4L7D3 A0A0L0CCV2 B4IKQ3 Q9V490 Q9XZC2 A0A0J9S132 A0A1W4VHQ6 B3P9Q4 A8VFE9 A0A0R3P0G3 A0A3B0K3Y7 B3N1J5 A0A286XBI8 Q38L56 G1T0K0 A0A287D9N3 A0A2K6CL21 A0A2K6L4C8 G1P774 A0A2I2ZFV9 A0A287AAF3 A0A2K5Y8D0 A0A2R9CK82 A0A2K5MMA0 H0XKU3 A0A2K5JGR6 H9FV32 A0A2K6S284 A0A2K6GSJ3 A0A3Q7XWK9 A0A2Y9D899 A0A2J8WG99 A0A2U3WCG5 I7G9J6 A0A2J8P422 F6S4R0 Q66SS1 P63015 P26367 A0A0Q9WKW4 B3KQG1 A0A1S3FEX4 P63016 G3TZY7 A0A2K5QU00 A0A3Q7U5D5 W5Q3U9 Q1LZF1 A0A341C1H6 A0A2Y9F604 A0A340XHU6 A0A384A1M6 A0A3Q0D361 H0WX77 A0A1U7S5R9 A0A0Q9XFH6 K7G268 A0A1V4JP70 A4GG27 G1LK37 D3DQZ8 A0A401SVE9 G1KAP0 A0A401P0Q4 A0A2K5Y8E0 L0MPR7 Q9I877 A0A0J9USS3
Pubmed
26354079
22118469
18362917
19820115
20566863
20798317
+ More
26823975 27289101 20075255 24845553 24495485 15632085 17994087 18057021 20479145 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10198632 22936249 21993624 19347868 22722832 25319552 17194215 16136131 25243066 1687460 11779807 16141072 15489334 1685142 1612585 1684639 9163426 17901057 18628401 21084637 1684738 1345175 17974005 16554811 7958875 7668281 18669648 19414065 20068231 24290376 10346815 9482572 8364574 8162071 8640214 9147640 9281415 9792406 9538891 9856761 10441571 10234503 9931324 10955655 10737978 11553050 11309364 11826019 12721955 12634864 12552561 16493447 17595013 21850189 24033328 16303743 24952136 7981749 11880342 24114637 20809919 17381049 17525177 20010809 11181995 30297745
26823975 27289101 20075255 24845553 24495485 15632085 17994087 18057021 20479145 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10198632 22936249 21993624 19347868 22722832 25319552 17194215 16136131 25243066 1687460 11779807 16141072 15489334 1685142 1612585 1684639 9163426 17901057 18628401 21084637 1684738 1345175 17974005 16554811 7958875 7668281 18669648 19414065 20068231 24290376 10346815 9482572 8364574 8162071 8640214 9147640 9281415 9792406 9538891 9856761 10441571 10234503 9931324 10955655 10737978 11553050 11309364 11826019 12721955 12634864 12552561 16493447 17595013 21850189 24033328 16303743 24952136 7981749 11880342 24114637 20809919 17381049 17525177 20010809 11181995 30297745
EMBL
KQ459585
KPI98175.1
KQ460627
KPJ13446.1
RSAL01000016
RVE53029.1
+ More
AGBW02007765 OWR54587.1 KQ971338 EFA02830.2 KYB27953.1 NEVH01020341 PNF21538.1 GEDC01015648 JAS21650.1 GECU01029138 JAS78568.1 UFQS01001062 UFQT01001062 SSX08780.1 SSX28692.1 UFQT01000002 SSX17089.1 DS235063 EEB11187.1 CVRI01000005 CRK87764.1 GL446691 EFN87393.1 GDHC01013583 JAQ05046.1 LJIJ01000173 ODN01076.1 AAZX01005026 AAZX01014109 AAZX01018084 AAZX01021306 KQ977622 KYN01346.1 KK852485 KDR22799.1 LNIX01000006 OXA52716.1 JH431363 GDIQ01020228 JAN74509.1 GAMC01003100 JAC03456.1 GAMC01003098 JAC03458.1 LN624822 CEH19758.1 OUUW01000017 SPP88948.1 CH475404 EDY70832.1 CH479285 EDW25268.1 CH916380 EDV90908.1 CH964272 EDW83579.2 CM000831 CH940665 ACY70471.1 EDW71088.1 CH933813 EDW10927.1 JRES01000552 KNC30268.1 CH480855 EDW52643.1 AE014135 AY051487 AAF59395.4 AAK92911.1 AF134350 AAD31712.1 CM002916 KMZ07347.1 CH954184 EDV45217.1 EU169112 ABW23132.1 KRT05135.1 SPP88947.1 CH902659 EDV33930.1 AAKN02048405 DQ217589 ABA90484.1 AAGW02037526 AAGW02037527 AAGW02037528 AGTP01048264 AAPE02023618 CABD030078140 AEMK02000009 AJFE02107122 AJFE02107123 AAQR03002365 JU334738 AFE78491.1 NDHI03003391 PNJ68803.1 PNJ68805.1 PNJ68816.1 PNJ68817.1 PNJ68824.1 PNJ68831.1 PNJ68838.1 AQIA01020593 AB171050 BAE88113.1 AACZ04016470 NBAG03000219 PNI78767.1 PNI78769.1 PNI78780.1 PNI78781.1 PNI78788.1 PNI78795.1 PNI78802.1 GAMS01006506 GAMR01007056 JAB16630.1 JAB26876.1 AY707088 KT580798 KT580799 AAU12168.1 ALU11231.1 X63963 Y19196 Y19199 AK028059 AK139054 BC011272 BC036957 M77842 M77844 M93650 AY047583 BX640762 Z83307 Z95332 BC011953 KRF85331.1 AK074881 BAG52023.1 U69644 AJ627631 AY540905 AY540906 BC128741 S74393 AMGL01032799 AMGL01032800 AMGL01032801 AMGL01032802 BC116038 KRG07295.1 AGCU01126135 AGCU01126136 AGCU01126137 LSYS01006880 OPJ73971.1 DQ870843 ABI98848.1 ACTA01025499 ACTA01033499 ACTA01041498 ACTA01049498 CH471064 EAW68234.1 BEZZ01000593 GCC34394.1 BFAA01002958 GCB66691.1 AGB96570.1 AF154555 AF154556 AAF73272.1 KMZ07346.1
AGBW02007765 OWR54587.1 KQ971338 EFA02830.2 KYB27953.1 NEVH01020341 PNF21538.1 GEDC01015648 JAS21650.1 GECU01029138 JAS78568.1 UFQS01001062 UFQT01001062 SSX08780.1 SSX28692.1 UFQT01000002 SSX17089.1 DS235063 EEB11187.1 CVRI01000005 CRK87764.1 GL446691 EFN87393.1 GDHC01013583 JAQ05046.1 LJIJ01000173 ODN01076.1 AAZX01005026 AAZX01014109 AAZX01018084 AAZX01021306 KQ977622 KYN01346.1 KK852485 KDR22799.1 LNIX01000006 OXA52716.1 JH431363 GDIQ01020228 JAN74509.1 GAMC01003100 JAC03456.1 GAMC01003098 JAC03458.1 LN624822 CEH19758.1 OUUW01000017 SPP88948.1 CH475404 EDY70832.1 CH479285 EDW25268.1 CH916380 EDV90908.1 CH964272 EDW83579.2 CM000831 CH940665 ACY70471.1 EDW71088.1 CH933813 EDW10927.1 JRES01000552 KNC30268.1 CH480855 EDW52643.1 AE014135 AY051487 AAF59395.4 AAK92911.1 AF134350 AAD31712.1 CM002916 KMZ07347.1 CH954184 EDV45217.1 EU169112 ABW23132.1 KRT05135.1 SPP88947.1 CH902659 EDV33930.1 AAKN02048405 DQ217589 ABA90484.1 AAGW02037526 AAGW02037527 AAGW02037528 AGTP01048264 AAPE02023618 CABD030078140 AEMK02000009 AJFE02107122 AJFE02107123 AAQR03002365 JU334738 AFE78491.1 NDHI03003391 PNJ68803.1 PNJ68805.1 PNJ68816.1 PNJ68817.1 PNJ68824.1 PNJ68831.1 PNJ68838.1 AQIA01020593 AB171050 BAE88113.1 AACZ04016470 NBAG03000219 PNI78767.1 PNI78769.1 PNI78780.1 PNI78781.1 PNI78788.1 PNI78795.1 PNI78802.1 GAMS01006506 GAMR01007056 JAB16630.1 JAB26876.1 AY707088 KT580798 KT580799 AAU12168.1 ALU11231.1 X63963 Y19196 Y19199 AK028059 AK139054 BC011272 BC036957 M77842 M77844 M93650 AY047583 BX640762 Z83307 Z95332 BC011953 KRF85331.1 AK074881 BAG52023.1 U69644 AJ627631 AY540905 AY540906 BC128741 S74393 AMGL01032799 AMGL01032800 AMGL01032801 AMGL01032802 BC116038 KRG07295.1 AGCU01126135 AGCU01126136 AGCU01126137 LSYS01006880 OPJ73971.1 DQ870843 ABI98848.1 ACTA01025499 ACTA01033499 ACTA01041498 ACTA01049498 CH471064 EAW68234.1 BEZZ01000593 GCC34394.1 BFAA01002958 GCB66691.1 AGB96570.1 AF154555 AF154556 AAF73272.1 KMZ07346.1
Proteomes
UP000053268
UP000053240
UP000283053
UP000007151
UP000007266
UP000235965
+ More
UP000009046 UP000183832 UP000008237 UP000094527 UP000002358 UP000078542 UP000027135 UP000198287 UP000268350 UP000001819 UP000008744 UP000001070 UP000007798 UP000008792 UP000009192 UP000037069 UP000001292 UP000000803 UP000192221 UP000008711 UP000007801 UP000005447 UP000001811 UP000005215 UP000233120 UP000233180 UP000001074 UP000001519 UP000008227 UP000233140 UP000240080 UP000233060 UP000005225 UP000233080 UP000233220 UP000233160 UP000286642 UP000248480 UP000245340 UP000233100 UP000002277 UP000008225 UP000000589 UP000005640 UP000081671 UP000002494 UP000007646 UP000233040 UP000286640 UP000002356 UP000009136 UP000252040 UP000248484 UP000265300 UP000261681 UP000189706 UP000189705 UP000007267 UP000190648 UP000008912 UP000287033 UP000001646 UP000288216
UP000009046 UP000183832 UP000008237 UP000094527 UP000002358 UP000078542 UP000027135 UP000198287 UP000268350 UP000001819 UP000008744 UP000001070 UP000007798 UP000008792 UP000009192 UP000037069 UP000001292 UP000000803 UP000192221 UP000008711 UP000007801 UP000005447 UP000001811 UP000005215 UP000233120 UP000233180 UP000001074 UP000001519 UP000008227 UP000233140 UP000240080 UP000233060 UP000005225 UP000233080 UP000233220 UP000233160 UP000286642 UP000248480 UP000245340 UP000233100 UP000002277 UP000008225 UP000000589 UP000005640 UP000081671 UP000002494 UP000007646 UP000233040 UP000286640 UP000002356 UP000009136 UP000252040 UP000248484 UP000265300 UP000261681 UP000189706 UP000189705 UP000007267 UP000190648 UP000008912 UP000287033 UP000001646 UP000288216
Interpro
SUPFAM
SSF46689
SSF46689
Gene 3D
ProteinModelPortal
A0A194Q439
A0A194RC13
A0A3S2LG24
A0A212FLI3
D1ZZR4
A0A139WJ10
+ More
A0A2J7PYX8 A0A1B6D7L7 A0A1B6HV64 A0A336L552 A0A336LKB8 E0VCT1 A0A1J1HK27 E2BA95 A0A146LD26 A0A1D2N7X8 K7J6S2 A0A195CKV0 A0A067RSK7 A0A226E7T7 T1JIT2 A0A0N8EJN0 W8BQD7 W8BW79 A0A0F7TDH4 A0A3B0KXY6 B5DRZ9 B4HC93 B4JZP2 B4NHL0 B4MEY7 B4L7D3 A0A0L0CCV2 B4IKQ3 Q9V490 Q9XZC2 A0A0J9S132 A0A1W4VHQ6 B3P9Q4 A8VFE9 A0A0R3P0G3 A0A3B0K3Y7 B3N1J5 A0A286XBI8 Q38L56 G1T0K0 A0A287D9N3 A0A2K6CL21 A0A2K6L4C8 G1P774 A0A2I2ZFV9 A0A287AAF3 A0A2K5Y8D0 A0A2R9CK82 A0A2K5MMA0 H0XKU3 A0A2K5JGR6 H9FV32 A0A2K6S284 A0A2K6GSJ3 A0A3Q7XWK9 A0A2Y9D899 A0A2J8WG99 A0A2U3WCG5 I7G9J6 A0A2J8P422 F6S4R0 Q66SS1 P63015 P26367 A0A0Q9WKW4 B3KQG1 A0A1S3FEX4 P63016 G3TZY7 A0A2K5QU00 A0A3Q7U5D5 W5Q3U9 Q1LZF1 A0A341C1H6 A0A2Y9F604 A0A340XHU6 A0A384A1M6 A0A3Q0D361 H0WX77 A0A1U7S5R9 A0A0Q9XFH6 K7G268 A0A1V4JP70 A4GG27 G1LK37 D3DQZ8 A0A401SVE9 G1KAP0 A0A401P0Q4 A0A2K5Y8E0 L0MPR7 Q9I877 A0A0J9USS3
A0A2J7PYX8 A0A1B6D7L7 A0A1B6HV64 A0A336L552 A0A336LKB8 E0VCT1 A0A1J1HK27 E2BA95 A0A146LD26 A0A1D2N7X8 K7J6S2 A0A195CKV0 A0A067RSK7 A0A226E7T7 T1JIT2 A0A0N8EJN0 W8BQD7 W8BW79 A0A0F7TDH4 A0A3B0KXY6 B5DRZ9 B4HC93 B4JZP2 B4NHL0 B4MEY7 B4L7D3 A0A0L0CCV2 B4IKQ3 Q9V490 Q9XZC2 A0A0J9S132 A0A1W4VHQ6 B3P9Q4 A8VFE9 A0A0R3P0G3 A0A3B0K3Y7 B3N1J5 A0A286XBI8 Q38L56 G1T0K0 A0A287D9N3 A0A2K6CL21 A0A2K6L4C8 G1P774 A0A2I2ZFV9 A0A287AAF3 A0A2K5Y8D0 A0A2R9CK82 A0A2K5MMA0 H0XKU3 A0A2K5JGR6 H9FV32 A0A2K6S284 A0A2K6GSJ3 A0A3Q7XWK9 A0A2Y9D899 A0A2J8WG99 A0A2U3WCG5 I7G9J6 A0A2J8P422 F6S4R0 Q66SS1 P63015 P26367 A0A0Q9WKW4 B3KQG1 A0A1S3FEX4 P63016 G3TZY7 A0A2K5QU00 A0A3Q7U5D5 W5Q3U9 Q1LZF1 A0A341C1H6 A0A2Y9F604 A0A340XHU6 A0A384A1M6 A0A3Q0D361 H0WX77 A0A1U7S5R9 A0A0Q9XFH6 K7G268 A0A1V4JP70 A4GG27 G1LK37 D3DQZ8 A0A401SVE9 G1KAP0 A0A401P0Q4 A0A2K5Y8E0 L0MPR7 Q9I877 A0A0J9USS3
PDB
6PAX
E-value=2.43752e-60,
Score=589
Ontologies
GO
GO:0007275
GO:0043565
GO:0006355
GO:0005634
GO:0003677
GO:0000977
GO:0000981
GO:0048854
GO:0045944
GO:0016319
GO:0008056
GO:0006357
GO:0048749
GO:0010628
GO:0035214
GO:0021778
GO:0019901
GO:0005654
GO:0030858
GO:0060041
GO:0021902
GO:0007435
GO:0005829
GO:0070410
GO:0070412
GO:0002052
GO:0035035
GO:0000790
GO:2000178
GO:0021986
GO:0048708
GO:0031625
GO:0030334
GO:0001764
GO:0006366
GO:0021912
GO:0021983
GO:0033365
GO:0000978
GO:0001228
GO:0000132
GO:0050680
GO:0021918
GO:0061303
GO:0042593
GO:0001227
GO:0023019
GO:1904798
GO:0021913
GO:0007224
GO:0001568
GO:0003309
GO:0071837
GO:0048505
GO:0001933
GO:0021796
GO:0009611
GO:0000979
GO:0008134
GO:0003322
GO:0001709
GO:1990830
GO:0045665
GO:0007411
GO:0021798
GO:0048596
GO:0030216
GO:0009786
GO:0009950
GO:0032808
GO:0021905
GO:0003682
GO:0042462
GO:0061072
GO:0002088
GO:0045893
GO:0009953
GO:0009952
GO:0022027
GO:0044212
GO:1901142
GO:0070094
GO:0021978
GO:0005737
GO:0021772
GO:0042660
GO:0002064
GO:0061034
GO:0007420
GO:2001224
GO:0045664
GO:0021543
GO:0030900
GO:0098598
GO:0043010
GO:0050767
GO:0120008
GO:0030154
GO:0010942
GO:0008285
GO:0001755
GO:0045165
GO:1904937
GO:0021593
GO:0061351
GO:0003310
GO:0010468
GO:0005667
GO:0007409
GO:0003700
GO:0003680
GO:0003690
GO:0030902
GO:0010975
GO:0003002
GO:0001654
GO:0048663
GO:0009887
GO:0007601
GO:0050768
GO:0007417
GO:0071333
GO:0021510
GO:0021549
GO:0071380
GO:0032869
GO:0044344
GO:0045471
GO:0035690
GO:0004842
GO:0016021
GO:0003917
GO:0016887
GO:0030145
GO:0006412
GO:0016876
GO:0043039
Topology
Subcellular location
Nucleus
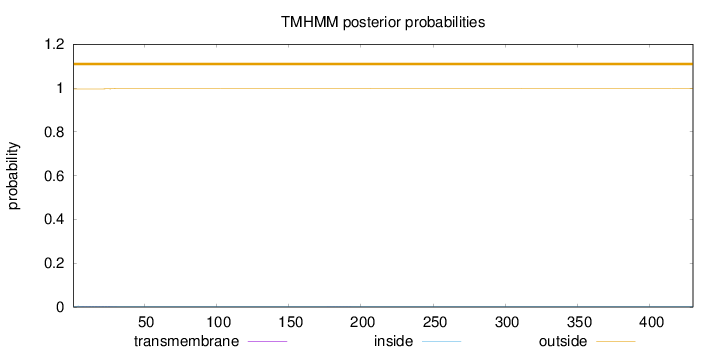
Length:
430
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0475600000000001
Exp number, first 60 AAs:
0.04327
Total prob of N-in:
0.00385
outside
1 - 430
Population Genetic Test Statistics
Pi
223.859866
Theta
171.907535
Tajima's D
0.805001
CLR
0.179202
CSRT
0.605119744012799
Interpretation
Uncertain
