Gene
KWMTBOMO08282
Annotation
PREDICTED:_transmembrane_7_superfamily_member_3-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.98
Sequence
CDS
ATGTTACTCAGCCAGTGCATTGACTTAATAATCATATTGTTATTTTTATCGTTTGTGTCCGTCTCTTCAATTGAAAGCATGGAGCAAATGACCATTGAACTCAAAGAACCAAAAACAGATGATACCAAAGACGTTTACAATGGCTTTATATACTTAAACCATTCATCTATAATACAAATAAACTTAAACAATTCAATCGAGGAGTTATCATTCATAATATTCCAAGTTCATAGTCATTTGTATAAAGTTACAATGTATAGAAATAAATTTCAAAGTGATACTTATGAATATGGAACTAATATTGGCCTGTACAGTAAAATAGCCGAAAATGATGTTTTCTATATTAAGAACATGAATAATGATATAGATTTAAAACTATATCTTGCAATACAAGGATATAAGAAACAGGATCCTGTACCCGGTGGGTGCAATATGGAGTCTCATGTTCCTGTCTCAACTGATCTTGTAATACAAGAGTTCTCAGATTACCTTTATGTTGATATCGCACCGGCACAAAACCCCCAAGAGCCAAATTGCTTAAATCCTTCCCAAGTTTCAGTCAAAGTCTATCTTATGTATATGCCCGAACGTAACTTTGATGCTGATGTGTACTTTGAAAGCATCAAAAGTATGATGAACTTGCCAAGCATTGAGAAAAATGGAGTTATTGTGCCCGCAATGTCGAAGATGATGCAAATGCGTCGTATCGTCGGCCTGTATCCCGGGACGGGTATGGTGTTCGTTGCCGTAGCCAAAGCCGGCGCCGACGGCTATGCCCTCTACGTGCCAGCCCAGACGTATGGATGTTCACCGTTTTCAAATGACGGATGCGAACTTATTAAGGACCTGTTGTCGTGCGTACTATGCGCGCTGCTTGTGTTCGTCGGACTGTTTCTCTGTTATTTACGTCATCGTTTCTTCAAGATCGAACTGTTCACGTTTGGATTCACAAGCGGATTTGTACTGACTTACATCTTTATATCATACCTCGTCGTTACCGATCGAACTGCCGTGGTAGCGGCTTCGGCGTTATCAGGTTTGTTCTTCGGCTCGATGTGGCTGATGTTCTGGTGGGTATACGGTATACCGATCATCGCCCTGCTGTTGACGTCATTGAATCTTGGATTCCTTATGGCATCACTGTTCTATTACGTCATACCAGATGACTTGTATTTCCGGGAGGACCTCAATTACTGGTTAATATTCGCATTAATCGTATTGGTGTGCGCGATAATAATGATAAGCGCGACCTTCGAAGCGAACATACTGGCCTGCGCGATACTCGGCGGTTACGCGGTTATATACCCTATAGACTACTACATTGGATCGAATTTGAAGTACATACTGATCAACACGATGAGACGGGCCACGGTGCCGAATTTCAAGAGTGCGACGCTCGTACTCAATTATCAATACAAAGATTTAGTACTGACACTCCTATGGGCATCGCTTACCGTTACAAGTTTCGCCTACCAATACTACACTAATTACGGCCGACCTCCCTTCCCCCCTCCACCTAGGGGCATCAGAGCTAGACCCCCGTAA
Protein
MLLSQCIDLIIILLFLSFVSVSSIESMEQMTIELKEPKTDDTKDVYNGFIYLNHSSIIQINLNNSIEELSFIIFQVHSHLYKVTMYRNKFQSDTYEYGTNIGLYSKIAENDVFYIKNMNNDIDLKLYLAIQGYKKQDPVPGGCNMESHVPVSTDLVIQEFSDYLYVDIAPAQNPQEPNCLNPSQVSVKVYLMYMPERNFDADVYFESIKSMMNLPSIEKNGVIVPAMSKMMQMRRIVGLYPGTGMVFVAVAKAGADGYALYVPAQTYGCSPFSNDGCELIKDLLSCVLCALLVFVGLFLCYLRHRFFKIELFTFGFTSGFVLTYIFISYLVVTDRTAVVAASALSGLFFGSMWLMFWWVYGIPIIALLLTSLNLGFLMASLFYYVIPDDLYFREDLNYWLIFALIVLVCAIIMISATFEANILACAILGGYAVIYPIDYYIGSNLKYILINTMRRATVPNFKSATLVLNYQYKDLVLTLLWASLTVTSFAYQYYTNYGRPPFPPPPRGIRARPP
Summary
Uniprot
A0A212FLL6
A0A2A4J1W6
A0A1E1WUM7
A0A3S2NZE3
A0A2H1VPH8
A0A194R7F1
+ More
A0A2A4J1C3 A0A2P8XPB7 A0A067RIE7 A0A2J7PYX0 A0A1B6KBX2 A0A1B6EXX6 A0A1B6EP84 A0A0C9RKG7 D1ZZR3 A0A1Y1M4B9 A0A0P5RIZ2 T1IRJ0 K7J6S3 A0A0P5KHD0 A0A0P5JX26 A0A0N8BX10 A0A0P5TKG9 A0A0P5JJ64 A0A0P5GCR8 A0A0P6EST1 A0A0N7ZRW9 A0A0P6EH48 A0A0P6GA52 A0A0N8CUA6 A0A0P6EHB0 A0A0N8E3F7 E9GXN3 A0A0P5PU83 A0A0P5QPW9 A0A0P6F483 A0A0P5ZFY3 A0A0P5R4V3 A0A0P4Z2V5 A0A0P5M480 A0A0P6EC48 A0A0P4YBG1 A0A0P6C276 A0A0P6EDT0 A0A0P5MWR3 A0A0N8D182 A0A0P6D2X9 A0A224XMG7 A0A0N8BF25 A0A226E767 T1I0B6 A0A1S3KEQ5 A0A0V0G520 A0A087ZRL1 A0A0P5A847 A0A1D2N751 A0A0P6IM40 A0A0M8ZWW5 A0A0P5L3P8 A0A0P5KFM9 A0A0P5LCZ8 C3Y542 G1KM35 A0A2R5LBG6 V9KEE9 L7MKV5 A0A444UDZ9 A0A131XMS0 G3W8A9 A0A131YM17 A0A151JXG2 A0A147BS09 A0A131XES6 E1ZYS6 K7FIS5 A0A443QZQ8 A0A151WPP0 R0JHS1 A0A224Z6S6 A0A452I860 U3IKQ8 A0A401S522 A0A3Q1MR19 L8I7L9 Q08DX3 W5NDL4
A0A2A4J1C3 A0A2P8XPB7 A0A067RIE7 A0A2J7PYX0 A0A1B6KBX2 A0A1B6EXX6 A0A1B6EP84 A0A0C9RKG7 D1ZZR3 A0A1Y1M4B9 A0A0P5RIZ2 T1IRJ0 K7J6S3 A0A0P5KHD0 A0A0P5JX26 A0A0N8BX10 A0A0P5TKG9 A0A0P5JJ64 A0A0P5GCR8 A0A0P6EST1 A0A0N7ZRW9 A0A0P6EH48 A0A0P6GA52 A0A0N8CUA6 A0A0P6EHB0 A0A0N8E3F7 E9GXN3 A0A0P5PU83 A0A0P5QPW9 A0A0P6F483 A0A0P5ZFY3 A0A0P5R4V3 A0A0P4Z2V5 A0A0P5M480 A0A0P6EC48 A0A0P4YBG1 A0A0P6C276 A0A0P6EDT0 A0A0P5MWR3 A0A0N8D182 A0A0P6D2X9 A0A224XMG7 A0A0N8BF25 A0A226E767 T1I0B6 A0A1S3KEQ5 A0A0V0G520 A0A087ZRL1 A0A0P5A847 A0A1D2N751 A0A0P6IM40 A0A0M8ZWW5 A0A0P5L3P8 A0A0P5KFM9 A0A0P5LCZ8 C3Y542 G1KM35 A0A2R5LBG6 V9KEE9 L7MKV5 A0A444UDZ9 A0A131XMS0 G3W8A9 A0A131YM17 A0A151JXG2 A0A147BS09 A0A131XES6 E1ZYS6 K7FIS5 A0A443QZQ8 A0A151WPP0 R0JHS1 A0A224Z6S6 A0A452I860 U3IKQ8 A0A401S522 A0A3Q1MR19 L8I7L9 Q08DX3 W5NDL4
Pubmed
EMBL
AGBW02007765
OWR54590.1
NWSH01003931
PCG65668.1
GDQN01000321
JAT90733.1
+ More
RSAL01000016 RVE53028.1 ODYU01003672 SOQ42739.1 KQ460627 KPJ13444.1 PCG65669.1 PYGN01001590 PSN33848.1 KK852485 KDR22798.1 NEVH01020341 PNF21547.1 GEBQ01032083 GEBQ01031062 GEBQ01016198 GEBQ01014015 GEBQ01013379 GEBQ01011668 GEBQ01008761 GEBQ01008214 JAT07894.1 JAT08915.1 JAT23779.1 JAT25962.1 JAT26598.1 JAT28309.1 JAT31216.1 JAT31763.1 GECZ01027088 JAS42681.1 GECZ01030045 JAS39724.1 GBYB01007371 JAG77138.1 KQ971338 EFA02406.2 GEZM01040964 GEZM01040963 JAV80684.1 GDIQ01119276 JAL32450.1 JH431363 AAZX01013962 GDIQ01184060 JAK67665.1 GDIQ01194061 JAK57664.1 GDIQ01136579 JAL15147.1 GDIP01130542 LRGB01001036 JAL73172.1 KZS13637.1 GDIQ01198204 JAK53521.1 GDIQ01246496 JAK05229.1 GDIQ01058457 JAN36280.1 GDIP01218585 JAJ04817.1 GDIQ01062654 JAN32083.1 GDIQ01039017 JAN55720.1 GDIP01098199 JAM05516.1 GDIQ01062653 JAN32084.1 GDIQ01065612 JAN29125.1 GL732573 EFX75690.1 GDIQ01144281 JAL07445.1 GDIQ01114036 JAL37690.1 GDIQ01053970 JAN40767.1 GDIP01044364 JAM59351.1 GDIQ01125383 JAL26343.1 GDIP01218586 JAJ04816.1 GDIQ01161071 JAK90654.1 GDIQ01065610 JAN29127.1 GDIP01229926 JAI93475.1 GDIP01007133 JAM96582.1 GDIQ01064161 JAN30576.1 GDIQ01150084 JAL01642.1 GDIP01078791 JAM24924.1 GDIQ01083544 JAN11193.1 GFTR01007093 JAW09333.1 GDIQ01184059 JAK67666.1 LNIX01000006 OXA52721.1 ACPB03011924 GECL01003650 JAP02474.1 GDIP01202788 JAJ20614.1 LJIJ01000173 ODN01077.1 GDIQ01004430 JAN90307.1 KQ435844 KOX71229.1 GDIQ01175904 JAK75821.1 GDIQ01191460 JAK60265.1 GDIQ01175905 JAK75820.1 GG666487 EEN64575.1 GGLE01002682 MBY06808.1 JW863609 AFO96126.1 GACK01001116 JAA63918.1 SCEB01214758 RXM33400.1 GEFM01007022 JAP68774.1 AEFK01194321 GEDV01009077 JAP79480.1 KQ981567 KYN39808.1 GEGO01001844 JAR93560.1 GEFH01003973 JAP64608.1 GL435242 EFN73616.1 AGCU01116746 AGCU01116747 AGCU01116748 NCKU01002944 RWS08499.1 KQ982851 KYQ49834.1 KB743948 EOA96531.1 GFPF01010876 MAA22022.1 ADON01116722 BEZZ01000086 GCC25439.1 JH881970 ELR51524.1 BC123528 AAI23529.1 AHAT01025453
RSAL01000016 RVE53028.1 ODYU01003672 SOQ42739.1 KQ460627 KPJ13444.1 PCG65669.1 PYGN01001590 PSN33848.1 KK852485 KDR22798.1 NEVH01020341 PNF21547.1 GEBQ01032083 GEBQ01031062 GEBQ01016198 GEBQ01014015 GEBQ01013379 GEBQ01011668 GEBQ01008761 GEBQ01008214 JAT07894.1 JAT08915.1 JAT23779.1 JAT25962.1 JAT26598.1 JAT28309.1 JAT31216.1 JAT31763.1 GECZ01027088 JAS42681.1 GECZ01030045 JAS39724.1 GBYB01007371 JAG77138.1 KQ971338 EFA02406.2 GEZM01040964 GEZM01040963 JAV80684.1 GDIQ01119276 JAL32450.1 JH431363 AAZX01013962 GDIQ01184060 JAK67665.1 GDIQ01194061 JAK57664.1 GDIQ01136579 JAL15147.1 GDIP01130542 LRGB01001036 JAL73172.1 KZS13637.1 GDIQ01198204 JAK53521.1 GDIQ01246496 JAK05229.1 GDIQ01058457 JAN36280.1 GDIP01218585 JAJ04817.1 GDIQ01062654 JAN32083.1 GDIQ01039017 JAN55720.1 GDIP01098199 JAM05516.1 GDIQ01062653 JAN32084.1 GDIQ01065612 JAN29125.1 GL732573 EFX75690.1 GDIQ01144281 JAL07445.1 GDIQ01114036 JAL37690.1 GDIQ01053970 JAN40767.1 GDIP01044364 JAM59351.1 GDIQ01125383 JAL26343.1 GDIP01218586 JAJ04816.1 GDIQ01161071 JAK90654.1 GDIQ01065610 JAN29127.1 GDIP01229926 JAI93475.1 GDIP01007133 JAM96582.1 GDIQ01064161 JAN30576.1 GDIQ01150084 JAL01642.1 GDIP01078791 JAM24924.1 GDIQ01083544 JAN11193.1 GFTR01007093 JAW09333.1 GDIQ01184059 JAK67666.1 LNIX01000006 OXA52721.1 ACPB03011924 GECL01003650 JAP02474.1 GDIP01202788 JAJ20614.1 LJIJ01000173 ODN01077.1 GDIQ01004430 JAN90307.1 KQ435844 KOX71229.1 GDIQ01175904 JAK75821.1 GDIQ01191460 JAK60265.1 GDIQ01175905 JAK75820.1 GG666487 EEN64575.1 GGLE01002682 MBY06808.1 JW863609 AFO96126.1 GACK01001116 JAA63918.1 SCEB01214758 RXM33400.1 GEFM01007022 JAP68774.1 AEFK01194321 GEDV01009077 JAP79480.1 KQ981567 KYN39808.1 GEGO01001844 JAR93560.1 GEFH01003973 JAP64608.1 GL435242 EFN73616.1 AGCU01116746 AGCU01116747 AGCU01116748 NCKU01002944 RWS08499.1 KQ982851 KYQ49834.1 KB743948 EOA96531.1 GFPF01010876 MAA22022.1 ADON01116722 BEZZ01000086 GCC25439.1 JH881970 ELR51524.1 BC123528 AAI23529.1 AHAT01025453
Proteomes
UP000007151
UP000218220
UP000283053
UP000053240
UP000245037
UP000027135
+ More
UP000235965 UP000007266 UP000002358 UP000076858 UP000000305 UP000198287 UP000015103 UP000085678 UP000005203 UP000094527 UP000053105 UP000001554 UP000001646 UP000007648 UP000078541 UP000000311 UP000007267 UP000285301 UP000075809 UP000291020 UP000016666 UP000287033 UP000009136 UP000018468
UP000235965 UP000007266 UP000002358 UP000076858 UP000000305 UP000198287 UP000015103 UP000085678 UP000005203 UP000094527 UP000053105 UP000001554 UP000001646 UP000007648 UP000078541 UP000000311 UP000007267 UP000285301 UP000075809 UP000291020 UP000016666 UP000287033 UP000009136 UP000018468
Pfam
PF13886 DUF4203
Interpro
IPR025256
DUF4203
ProteinModelPortal
A0A212FLL6
A0A2A4J1W6
A0A1E1WUM7
A0A3S2NZE3
A0A2H1VPH8
A0A194R7F1
+ More
A0A2A4J1C3 A0A2P8XPB7 A0A067RIE7 A0A2J7PYX0 A0A1B6KBX2 A0A1B6EXX6 A0A1B6EP84 A0A0C9RKG7 D1ZZR3 A0A1Y1M4B9 A0A0P5RIZ2 T1IRJ0 K7J6S3 A0A0P5KHD0 A0A0P5JX26 A0A0N8BX10 A0A0P5TKG9 A0A0P5JJ64 A0A0P5GCR8 A0A0P6EST1 A0A0N7ZRW9 A0A0P6EH48 A0A0P6GA52 A0A0N8CUA6 A0A0P6EHB0 A0A0N8E3F7 E9GXN3 A0A0P5PU83 A0A0P5QPW9 A0A0P6F483 A0A0P5ZFY3 A0A0P5R4V3 A0A0P4Z2V5 A0A0P5M480 A0A0P6EC48 A0A0P4YBG1 A0A0P6C276 A0A0P6EDT0 A0A0P5MWR3 A0A0N8D182 A0A0P6D2X9 A0A224XMG7 A0A0N8BF25 A0A226E767 T1I0B6 A0A1S3KEQ5 A0A0V0G520 A0A087ZRL1 A0A0P5A847 A0A1D2N751 A0A0P6IM40 A0A0M8ZWW5 A0A0P5L3P8 A0A0P5KFM9 A0A0P5LCZ8 C3Y542 G1KM35 A0A2R5LBG6 V9KEE9 L7MKV5 A0A444UDZ9 A0A131XMS0 G3W8A9 A0A131YM17 A0A151JXG2 A0A147BS09 A0A131XES6 E1ZYS6 K7FIS5 A0A443QZQ8 A0A151WPP0 R0JHS1 A0A224Z6S6 A0A452I860 U3IKQ8 A0A401S522 A0A3Q1MR19 L8I7L9 Q08DX3 W5NDL4
A0A2A4J1C3 A0A2P8XPB7 A0A067RIE7 A0A2J7PYX0 A0A1B6KBX2 A0A1B6EXX6 A0A1B6EP84 A0A0C9RKG7 D1ZZR3 A0A1Y1M4B9 A0A0P5RIZ2 T1IRJ0 K7J6S3 A0A0P5KHD0 A0A0P5JX26 A0A0N8BX10 A0A0P5TKG9 A0A0P5JJ64 A0A0P5GCR8 A0A0P6EST1 A0A0N7ZRW9 A0A0P6EH48 A0A0P6GA52 A0A0N8CUA6 A0A0P6EHB0 A0A0N8E3F7 E9GXN3 A0A0P5PU83 A0A0P5QPW9 A0A0P6F483 A0A0P5ZFY3 A0A0P5R4V3 A0A0P4Z2V5 A0A0P5M480 A0A0P6EC48 A0A0P4YBG1 A0A0P6C276 A0A0P6EDT0 A0A0P5MWR3 A0A0N8D182 A0A0P6D2X9 A0A224XMG7 A0A0N8BF25 A0A226E767 T1I0B6 A0A1S3KEQ5 A0A0V0G520 A0A087ZRL1 A0A0P5A847 A0A1D2N751 A0A0P6IM40 A0A0M8ZWW5 A0A0P5L3P8 A0A0P5KFM9 A0A0P5LCZ8 C3Y542 G1KM35 A0A2R5LBG6 V9KEE9 L7MKV5 A0A444UDZ9 A0A131XMS0 G3W8A9 A0A131YM17 A0A151JXG2 A0A147BS09 A0A131XES6 E1ZYS6 K7FIS5 A0A443QZQ8 A0A151WPP0 R0JHS1 A0A224Z6S6 A0A452I860 U3IKQ8 A0A401S522 A0A3Q1MR19 L8I7L9 Q08DX3 W5NDL4
Ontologies
Topology
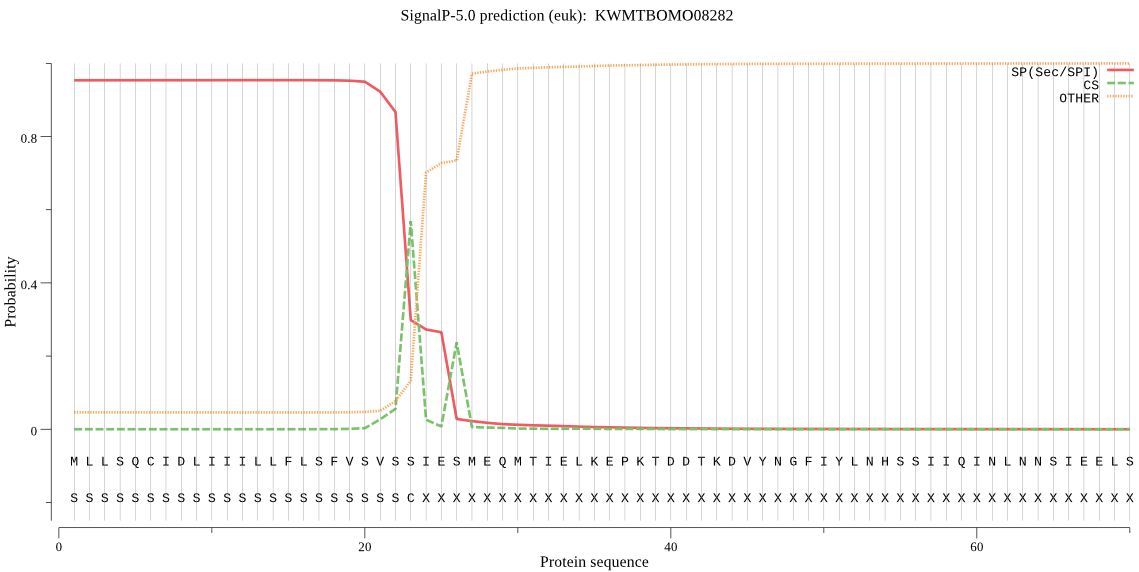
SignalP
Position: 1 - 23,
Likelihood: 0.954025
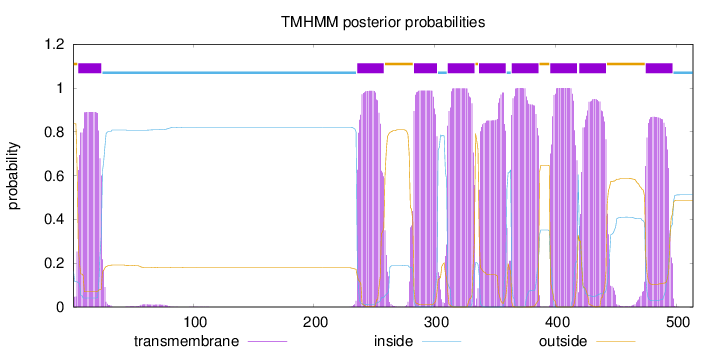
Length:
514
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
184.75534
Exp number, first 60 AAs:
17.44953
Total prob of N-in:
0.16254
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 24
inside
25 - 235
TMhelix
236 - 258
outside
259 - 282
TMhelix
283 - 302
inside
303 - 310
TMhelix
311 - 333
outside
334 - 336
TMhelix
337 - 359
inside
360 - 363
TMhelix
364 - 386
outside
387 - 395
TMhelix
396 - 418
inside
419 - 419
TMhelix
420 - 442
outside
443 - 474
TMhelix
475 - 497
inside
498 - 514
Population Genetic Test Statistics
Pi
183.65679
Theta
151.804091
Tajima's D
0.501984
CLR
0.272389
CSRT
0.511174441277936
Interpretation
Uncertain
