Gene
KWMTBOMO08270
Pre Gene Modal
BGIBMGA009440
Annotation
PREDICTED:_von_Willebrand_factor_A_domain-containing_protein_7_[Gekko_japonicus]
Location in the cell
Mitochondrial Reliability : 1.432 Nuclear Reliability : 1.147
Sequence
CDS
ATGCGTTTAATTCATTTCATATTACTCCTGTACTGTTTCGGAAATATACGAACGAAGAATGAGAGCGGCCGGAGTCTCACTTTTGTTATAGATCGCTCGTATTCACTGCAACTTCTAATAAAACAGTTCAAGGAAGCGTCATTGAATCTCACCAGTAAATTGATGGGACGTAAATCAAGTGATATCGTTAACGTAACACTTATTGTGTTCGATGATCCAACATACAAGCTTGTAGTGGACACCACGGACATTAGCGAGTTCGAGAAAAACTTAAAATCACTGGAAACTTACTACGACATTAAAAACAAAGATTGCCCTGAGATGAGTCTGTCTGCTCTCAAAATGGCTTTGACTAGGAGCTTGCCGAGATCATATATACTTCTATTCACCGACGCTACAGCAAAAGATGTATGGCTTTTGGACGAAGTTAAAATACTATCTAAGGAAAAGCAGAGCAAGGTAAGATCTATGTTGTAA
Protein
MRLIHFILLLYCFGNIRTKNESGRSLTFVIDRSYSLQLLIKQFKEASLNLTSKLMGRKSSDIVNVTLIVFDDPTYKLVVDTTDISEFEKNLKSLETYYDIKNKDCPEMSLSALKMALTRSLPRSYILLFTDATAKDVWLLDEVKILSKEKQSKVRSML
Summary
Uniprot
T1EJA1
G1KX23
T1EJ97
A0A044SXW9
A0A1S0UMB4
A0A1Y3E5W1
+ More
C3Z8Z3 A0A401SJ60 A0A158Q9J1 A0A3P7DZB1 A0A0V1DJB6 A0A0V1HBV0 A0A0V1HBR0 A0A0V1DI29 A0A0V1DI53 A0A183V948 A0A0V1FI13 A0A0V1FI33 A0A0V0WXG9 A0A0V0VTL5 A0A0V1KWX8 A0A0V1NX02 A0A0V0S0I9 A0A0V1BJK0 A0A0V1A256 A0A0V1DYW1 A0A0V1IE20 A0A0V1DY64 A0A0V0S0N1 A0A0V0S0T3 A0A0V1NWG3 A0A0V1BJH5 A0A0V1NWI7 A0A0V1BJ77 A0A0V1BK67 A0A0V0WXD9 A0A0V0VTJ3 A0A0V1KVV1 A0A0V0U7T3 A0A0V1A1V7 A0A0V0WXR1 A0A0V1MLA8 A0A0V0VU77 A0A0V1KVS9 A0A0V0U8D7 A0A0V1A1Z7 A0A0V0WXR7 A0A0V0U9F2 A0A0V1MLS9 A0A183ISW3 A0A0N1I874 C4A0G4 A0A158R5T7 A0A0R3QT82 A0A087U6Q6 H3ADQ7 A0A0N5CMP2 A0A0R3RPN4 A0A158Q3H3 A0A3B3X3L6 A0A0B8RXN5 A0A444TI42 A0A0K0JA35 A0A146WR83 A0A146WQT6 A0A1S3KCR7 A0A1S3IRE7 A0A3Q2Q6M6 A0A3B3UCJ0 A0A3S2PS86 A0A2I4B4G3 A0A2I4B4D6 E7EY71 A0A1U8DJP2 A0A0S7HR98 A0A3Q0CSE1 A0A3P6TKL1 A0A3P9P691 A0A3P9AE33 A0A1S3PI68 A0A1S3PI86 A0A3Q4GUD4 E7F3V2 M3ZDA5 K1R2P7 A0A1I8F0W4 A0A0N4TJ88 A0A3Q3C3Q9 A0A3P9BQE6 A0A3B4GRR4 A0A3P8PUT5 I3JXY4 A0A067QUP4 A0A3P8SNR4 A0A2U9BRP6 A0A1E1WM33 A0A060VWR8 A0A3B5MHR2 A0A3B4XL25
C3Z8Z3 A0A401SJ60 A0A158Q9J1 A0A3P7DZB1 A0A0V1DJB6 A0A0V1HBV0 A0A0V1HBR0 A0A0V1DI29 A0A0V1DI53 A0A183V948 A0A0V1FI13 A0A0V1FI33 A0A0V0WXG9 A0A0V0VTL5 A0A0V1KWX8 A0A0V1NX02 A0A0V0S0I9 A0A0V1BJK0 A0A0V1A256 A0A0V1DYW1 A0A0V1IE20 A0A0V1DY64 A0A0V0S0N1 A0A0V0S0T3 A0A0V1NWG3 A0A0V1BJH5 A0A0V1NWI7 A0A0V1BJ77 A0A0V1BK67 A0A0V0WXD9 A0A0V0VTJ3 A0A0V1KVV1 A0A0V0U7T3 A0A0V1A1V7 A0A0V0WXR1 A0A0V1MLA8 A0A0V0VU77 A0A0V1KVS9 A0A0V0U8D7 A0A0V1A1Z7 A0A0V0WXR7 A0A0V0U9F2 A0A0V1MLS9 A0A183ISW3 A0A0N1I874 C4A0G4 A0A158R5T7 A0A0R3QT82 A0A087U6Q6 H3ADQ7 A0A0N5CMP2 A0A0R3RPN4 A0A158Q3H3 A0A3B3X3L6 A0A0B8RXN5 A0A444TI42 A0A0K0JA35 A0A146WR83 A0A146WQT6 A0A1S3KCR7 A0A1S3IRE7 A0A3Q2Q6M6 A0A3B3UCJ0 A0A3S2PS86 A0A2I4B4G3 A0A2I4B4D6 E7EY71 A0A1U8DJP2 A0A0S7HR98 A0A3Q0CSE1 A0A3P6TKL1 A0A3P9P691 A0A3P9AE33 A0A1S3PI68 A0A1S3PI86 A0A3Q4GUD4 E7F3V2 M3ZDA5 K1R2P7 A0A1I8F0W4 A0A0N4TJ88 A0A3Q3C3Q9 A0A3P9BQE6 A0A3B4GRR4 A0A3P8PUT5 I3JXY4 A0A067QUP4 A0A3P8SNR4 A0A2U9BRP6 A0A1E1WM33 A0A060VWR8 A0A3B5MHR2 A0A3B4XL25
Pubmed
EMBL
AMQM01007465
KB097620
ESN93353.1
ESN93355.1
CMVM020000118
JH712067
+ More
EJD76713.1 LVZM01023376 OUC40030.1 GG666598 EEN50905.1 BEZZ01000303 GCC30457.1 UXUI01007290 VDD86792.1 UYWW01001006 VDM09757.1 JYDI01000002 KRY61123.1 JYDP01000098 KRZ07695.1 KRZ07696.1 KRY61125.1 KRY61124.1 UYWY01024289 VDM48587.1 JYDT01000085 KRY85688.1 KRY85687.1 JYDK01000041 KRX80459.1 JYDN01000009 KRX66814.1 JYDW01000225 KRZ51458.1 JYDM01000086 KRZ88323.1 JYDL01000051 KRX20240.1 JYDH01000036 KRY37114.1 JYDQ01000041 KRY18897.1 JYDR01000166 JYDS01000219 JYDV01000017 KRY66518.1 KRZ21053.1 KRZ42041.1 KRY66520.1 KRZ21051.1 KRZ42039.1 KRY66519.1 KRZ21050.1 KRZ42040.1 KRX20241.1 KRX20239.1 KRZ88324.1 KRY37111.1 KRZ88322.1 KRY37113.1 KRY37112.1 KRX80458.1 KRX66815.1 KRZ51460.1 JYDJ01000043 KRX47396.1 KRY18896.1 KRX80457.1 JYDO01000076 KRZ72617.1 KRX66813.1 KRZ51459.1 KRX47397.1 KRY18895.1 KRX80456.1 KRX47395.1 KRZ72618.1 UZAM01009981 VDP10636.1 KQ460426 KPJ14779.1 GG666804 EEN41706.1 UZAG01016692 VDO30100.1 KK118472 KFM73045.1 AFYH01199621 AFYH01199622 AFYH01199623 AFYH01199624 AFYH01199625 AFYH01199626 AFYH01199627 AFYH01199628 AFYH01199629 AFYH01199630 UYYF01000183 VDM96861.1 UYYG01001157 VDN56791.1 GBSH01001123 JAG67902.1 SAUD01002510 RXG69659.1 GCES01054796 JAR31527.1 GCES01054797 JAR31526.1 CM012445 RVE68435.1 BX548021 GBYX01437714 JAO43638.1 UYRX01000551 VDK83743.1 CR391979 CR457445 JH819016 EKC40013.1 UZAD01013133 VDN89500.1 AERX01013686 KK852989 KDR12788.1 CP026251 AWP06721.1 GDQN01003046 JAT88008.1 FR904268 CDQ56760.1
EJD76713.1 LVZM01023376 OUC40030.1 GG666598 EEN50905.1 BEZZ01000303 GCC30457.1 UXUI01007290 VDD86792.1 UYWW01001006 VDM09757.1 JYDI01000002 KRY61123.1 JYDP01000098 KRZ07695.1 KRZ07696.1 KRY61125.1 KRY61124.1 UYWY01024289 VDM48587.1 JYDT01000085 KRY85688.1 KRY85687.1 JYDK01000041 KRX80459.1 JYDN01000009 KRX66814.1 JYDW01000225 KRZ51458.1 JYDM01000086 KRZ88323.1 JYDL01000051 KRX20240.1 JYDH01000036 KRY37114.1 JYDQ01000041 KRY18897.1 JYDR01000166 JYDS01000219 JYDV01000017 KRY66518.1 KRZ21053.1 KRZ42041.1 KRY66520.1 KRZ21051.1 KRZ42039.1 KRY66519.1 KRZ21050.1 KRZ42040.1 KRX20241.1 KRX20239.1 KRZ88324.1 KRY37111.1 KRZ88322.1 KRY37113.1 KRY37112.1 KRX80458.1 KRX66815.1 KRZ51460.1 JYDJ01000043 KRX47396.1 KRY18896.1 KRX80457.1 JYDO01000076 KRZ72617.1 KRX66813.1 KRZ51459.1 KRX47397.1 KRY18895.1 KRX80456.1 KRX47395.1 KRZ72618.1 UZAM01009981 VDP10636.1 KQ460426 KPJ14779.1 GG666804 EEN41706.1 UZAG01016692 VDO30100.1 KK118472 KFM73045.1 AFYH01199621 AFYH01199622 AFYH01199623 AFYH01199624 AFYH01199625 AFYH01199626 AFYH01199627 AFYH01199628 AFYH01199629 AFYH01199630 UYYF01000183 VDM96861.1 UYYG01001157 VDN56791.1 GBSH01001123 JAG67902.1 SAUD01002510 RXG69659.1 GCES01054796 JAR31527.1 GCES01054797 JAR31526.1 CM012445 RVE68435.1 BX548021 GBYX01437714 JAO43638.1 UYRX01000551 VDK83743.1 CR391979 CR457445 JH819016 EKC40013.1 UZAD01013133 VDN89500.1 AERX01013686 KK852989 KDR12788.1 CP026251 AWP06721.1 GDQN01003046 JAT88008.1 FR904268 CDQ56760.1
Proteomes
UP000015101
UP000001646
UP000024404
UP000243006
UP000001554
UP000287033
+ More
UP000038041 UP000274131 UP000270924 UP000054653 UP000055024 UP000050794 UP000267007 UP000054995 UP000054673 UP000054681 UP000054721 UP000054924 UP000054630 UP000054776 UP000054783 UP000054632 UP000054805 UP000054826 UP000055048 UP000054843 UP000050793 UP000053240 UP000046393 UP000050602 UP000054359 UP000008672 UP000046394 UP000276776 UP000050640 UP000038040 UP000274756 UP000261480 UP000288706 UP000006672 UP000085678 UP000265000 UP000261500 UP000192220 UP000000437 UP000189705 UP000189706 UP000277928 UP000242638 UP000265140 UP000087266 UP000261580 UP000002852 UP000005408 UP000093561 UP000038020 UP000278627 UP000264840 UP000265160 UP000261460 UP000265100 UP000005207 UP000027135 UP000265080 UP000246464 UP000193380 UP000261380 UP000261360
UP000038041 UP000274131 UP000270924 UP000054653 UP000055024 UP000050794 UP000267007 UP000054995 UP000054673 UP000054681 UP000054721 UP000054924 UP000054630 UP000054776 UP000054783 UP000054632 UP000054805 UP000054826 UP000055048 UP000054843 UP000050793 UP000053240 UP000046393 UP000050602 UP000054359 UP000008672 UP000046394 UP000276776 UP000050640 UP000038040 UP000274756 UP000261480 UP000288706 UP000006672 UP000085678 UP000265000 UP000261500 UP000192220 UP000000437 UP000189705 UP000189706 UP000277928 UP000242638 UP000265140 UP000087266 UP000261580 UP000002852 UP000005408 UP000093561 UP000038020 UP000278627 UP000264840 UP000265160 UP000261460 UP000265100 UP000005207 UP000027135 UP000265080 UP000246464 UP000193380 UP000261380 UP000261360
Pfam
Interpro
IPR036465
vWFA_dom_sf
+ More
IPR013098 Ig_I-set
IPR001881 EGF-like_Ca-bd_dom
IPR026823 cEGF
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR003599 Ig_sub
IPR036179 Ig-like_dom_sf
IPR000742 EGF-like_dom
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR003598 Ig_sub2
IPR013106 Ig_V-set
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR013151 Immunoglobulin
IPR002035 VWF_A
IPR001604 DNA/RNA_non-sp_Endonuclease
IPR000884 TSP1_rpt
IPR009017 GFP
IPR036383 TSP1_rpt_sf
IPR006605 G2_nidogen/fibulin_G2F
IPR032984 Hemicentin-2
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018247 EF_Hand_1_Ca_BS
IPR013098 Ig_I-set
IPR001881 EGF-like_Ca-bd_dom
IPR026823 cEGF
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR003599 Ig_sub
IPR036179 Ig-like_dom_sf
IPR000742 EGF-like_dom
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR003598 Ig_sub2
IPR013106 Ig_V-set
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR013151 Immunoglobulin
IPR002035 VWF_A
IPR001604 DNA/RNA_non-sp_Endonuclease
IPR000884 TSP1_rpt
IPR009017 GFP
IPR036383 TSP1_rpt_sf
IPR006605 G2_nidogen/fibulin_G2F
IPR032984 Hemicentin-2
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018247 EF_Hand_1_Ca_BS
SUPFAM
Gene 3D
CDD
ProteinModelPortal
T1EJA1
G1KX23
T1EJ97
A0A044SXW9
A0A1S0UMB4
A0A1Y3E5W1
+ More
C3Z8Z3 A0A401SJ60 A0A158Q9J1 A0A3P7DZB1 A0A0V1DJB6 A0A0V1HBV0 A0A0V1HBR0 A0A0V1DI29 A0A0V1DI53 A0A183V948 A0A0V1FI13 A0A0V1FI33 A0A0V0WXG9 A0A0V0VTL5 A0A0V1KWX8 A0A0V1NX02 A0A0V0S0I9 A0A0V1BJK0 A0A0V1A256 A0A0V1DYW1 A0A0V1IE20 A0A0V1DY64 A0A0V0S0N1 A0A0V0S0T3 A0A0V1NWG3 A0A0V1BJH5 A0A0V1NWI7 A0A0V1BJ77 A0A0V1BK67 A0A0V0WXD9 A0A0V0VTJ3 A0A0V1KVV1 A0A0V0U7T3 A0A0V1A1V7 A0A0V0WXR1 A0A0V1MLA8 A0A0V0VU77 A0A0V1KVS9 A0A0V0U8D7 A0A0V1A1Z7 A0A0V0WXR7 A0A0V0U9F2 A0A0V1MLS9 A0A183ISW3 A0A0N1I874 C4A0G4 A0A158R5T7 A0A0R3QT82 A0A087U6Q6 H3ADQ7 A0A0N5CMP2 A0A0R3RPN4 A0A158Q3H3 A0A3B3X3L6 A0A0B8RXN5 A0A444TI42 A0A0K0JA35 A0A146WR83 A0A146WQT6 A0A1S3KCR7 A0A1S3IRE7 A0A3Q2Q6M6 A0A3B3UCJ0 A0A3S2PS86 A0A2I4B4G3 A0A2I4B4D6 E7EY71 A0A1U8DJP2 A0A0S7HR98 A0A3Q0CSE1 A0A3P6TKL1 A0A3P9P691 A0A3P9AE33 A0A1S3PI68 A0A1S3PI86 A0A3Q4GUD4 E7F3V2 M3ZDA5 K1R2P7 A0A1I8F0W4 A0A0N4TJ88 A0A3Q3C3Q9 A0A3P9BQE6 A0A3B4GRR4 A0A3P8PUT5 I3JXY4 A0A067QUP4 A0A3P8SNR4 A0A2U9BRP6 A0A1E1WM33 A0A060VWR8 A0A3B5MHR2 A0A3B4XL25
C3Z8Z3 A0A401SJ60 A0A158Q9J1 A0A3P7DZB1 A0A0V1DJB6 A0A0V1HBV0 A0A0V1HBR0 A0A0V1DI29 A0A0V1DI53 A0A183V948 A0A0V1FI13 A0A0V1FI33 A0A0V0WXG9 A0A0V0VTL5 A0A0V1KWX8 A0A0V1NX02 A0A0V0S0I9 A0A0V1BJK0 A0A0V1A256 A0A0V1DYW1 A0A0V1IE20 A0A0V1DY64 A0A0V0S0N1 A0A0V0S0T3 A0A0V1NWG3 A0A0V1BJH5 A0A0V1NWI7 A0A0V1BJ77 A0A0V1BK67 A0A0V0WXD9 A0A0V0VTJ3 A0A0V1KVV1 A0A0V0U7T3 A0A0V1A1V7 A0A0V0WXR1 A0A0V1MLA8 A0A0V0VU77 A0A0V1KVS9 A0A0V0U8D7 A0A0V1A1Z7 A0A0V0WXR7 A0A0V0U9F2 A0A0V1MLS9 A0A183ISW3 A0A0N1I874 C4A0G4 A0A158R5T7 A0A0R3QT82 A0A087U6Q6 H3ADQ7 A0A0N5CMP2 A0A0R3RPN4 A0A158Q3H3 A0A3B3X3L6 A0A0B8RXN5 A0A444TI42 A0A0K0JA35 A0A146WR83 A0A146WQT6 A0A1S3KCR7 A0A1S3IRE7 A0A3Q2Q6M6 A0A3B3UCJ0 A0A3S2PS86 A0A2I4B4G3 A0A2I4B4D6 E7EY71 A0A1U8DJP2 A0A0S7HR98 A0A3Q0CSE1 A0A3P6TKL1 A0A3P9P691 A0A3P9AE33 A0A1S3PI68 A0A1S3PI86 A0A3Q4GUD4 E7F3V2 M3ZDA5 K1R2P7 A0A1I8F0W4 A0A0N4TJ88 A0A3Q3C3Q9 A0A3P9BQE6 A0A3B4GRR4 A0A3P8PUT5 I3JXY4 A0A067QUP4 A0A3P8SNR4 A0A2U9BRP6 A0A1E1WM33 A0A060VWR8 A0A3B5MHR2 A0A3B4XL25
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.681960
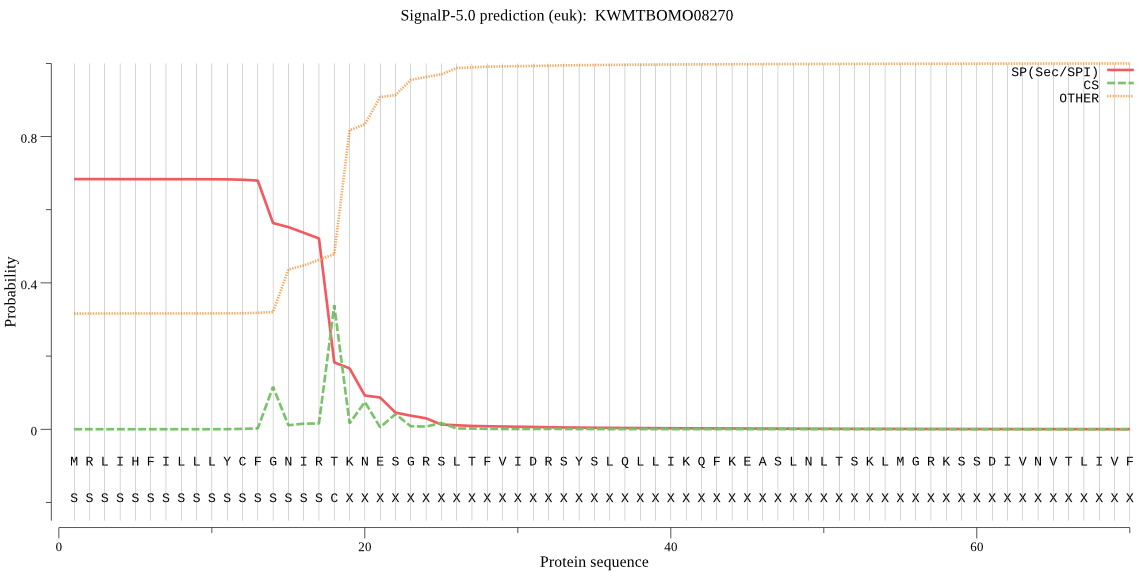
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00483
Exp number, first 60 AAs:
0.00244
Total prob of N-in:
0.01995
outside
1 - 158

Population Genetic Test Statistics
Pi
162.653183
Theta
147.907115
Tajima's D
0.313382
CLR
0
CSRT
0.463226838658067
Interpretation
Uncertain
