Gene
KWMTBOMO08264 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009442
Annotation
hypothetical_protein_KGM_12367_[Danaus_plexippus]
Full name
Hemicentin-1
Alternative Name
Fibulin-6
Location in the cell
Nuclear Reliability : 1.259 PlasmaMembrane Reliability : 1.451
Sequence
CDS
ATGTTCCGAATAGAACCGAAACAAGGGAAATATTGTATGTTCATTCGGACTCACGAGGTAGTTTCAGTAACGCTGTACATAAAAAGTGATTTGGTCTTCTGTTTCGGATTCTCACCTAAATACCCTAAATCATTGGAGGAGACCAGCACTAGGCCTATGGCAGGTCGTTCGAACTACATACTAATAAGCGTAGTGGACACAAAGATACGATTGGTTTCGTTGGAAATGAAATGTGGTAATATGACGACTGTCATAAGCTTTGAATATACCACTAACGAAAGGTATTACACGAGCAGAGCATTTGTGGATCCCAACAGATTATGTCGCATCACGGCAATAGGAGAGGATGAGAATTCTCAAGGATTTATAGGTGCTACAAAGTTACTCAAGCCACAAGAACAAGTGTCAAATATTTCGGCGCTAAAACCAACTGTAGTGATTATTGATCCAGAATATACGCTGTTCGACTTTGGCACCGATGTTTCATTGACGTGCAAAATAAAAGGACATCCAGAGCCTGTTGTGAAATGGGAAGATGATGACGGCCACTTACATCCTTCCGAGCTGATCCTGCTCGAATTACCCTCGACTTACATCAGTTACGTTACGATAACGAACATAACACGGAACATGACCATTTCCTGCAAAGCTGAGAACACAGAAGGTTCATCTGAAACCAGTCTCCATGTTTATATTAACAAGACATTTACTTTTGACATTGTGCAAACGCCGTCAGATATAACAATAGAATACGGCGAAGAAGATACAGTATTTTGTGAGGTGTCTGCTTACCCCGAAGCCACCACAAAATGGTATCACAACAACTCCGAAATAGTCTCCTCCCAGAATCTCCAGCTGATACCAGACGACAATGCCATGGTTATTAAAAACATGACTTTAGATTACACGGGCGATTATAAATGTGTCGTCGAGAACGTCGCTAATAAAAAAGAATTTGATTTTGTCATCAGTATATCCGGCTTGGAATCCCCTCAGATTGAAATCAATACACGAGAAATCGTATTGAGACCAGGTGATTCAGCGGAACTCAATTGCAGGATCTTGAAAGGAAATCCTGCCCCTCTTATCACTTGGCAACACAAATCTGCAGACGAATTCGCTTCTGACGAACTACCCGAATATTCTATTATAGTTGGAAATAAGCTTAGGATTTTATCTATTAAAAAGGAACATGAAGGAATATACCGATGCACAGCTGTAAATCTCATGGGCGAAAGCTCAGCGGAAGTAACGTTGAAAGTTCAATTTGCTCCTGTCATTAAAGATGATGTTATATATCCCCAAACTGTGCCGGTGAAGGAGGGAGATAATGTGGAACTACCTTGTGACGTCACAGCATCACCGGAAGCTGTAGTTAGATGGGAAATGTCACAAGATGATGTTATAATCCCACTGGATCAAAGACACGTTACAGATGACCAGAATACACATAGATTTACTGCCTTGTGGAGAGATTCCGGTCATTATCACTGTATTGCCGAAAATGCCCTGGGAACAGCAAAGAAAACCATTCTAGTAAATGTCTTAGTGGCCCCATACATCGAAACACCACAGTCAAAAACATTGACAGTTCGTTCAGGCTCAACTGTCAAACTAGCGTGCAACGTTCTATACGGTAATCCGGCTCCATCTTTGAAATGGAAATTCATAAATAAGGATTCCACGTCTAGGATACTCAGAAACAACAGGACGAGTTCGTTAACCTTGAACAATGTCAACAAATTAAATGAAGGTTCCTATATGTGCATCGTGGACAATGCACTAGGCTCTGACAGGATTAAATATCAATTAAAAGTTGAATAA
Protein
MFRIEPKQGKYCMFIRTHEVVSVTLYIKSDLVFCFGFSPKYPKSLEETSTRPMAGRSNYILISVVDTKIRLVSLEMKCGNMTTVISFEYTTNERYYTSRAFVDPNRLCRITAIGEDENSQGFIGATKLLKPQEQVSNISALKPTVVIIDPEYTLFDFGTDVSLTCKIKGHPEPVVKWEDDDGHLHPSELILLELPSTYISYVTITNITRNMTISCKAENTEGSSETSLHVYINKTFTFDIVQTPSDITIEYGEEDTVFCEVSAYPEATTKWYHNNSEIVSSQNLQLIPDDNAMVIKNMTLDYTGDYKCVVENVANKKEFDFVISISGLESPQIEINTREIVLRPGDSAELNCRILKGNPAPLITWQHKSADEFASDELPEYSIIVGNKLRILSIKKEHEGIYRCTAVNLMGESSAEVTLKVQFAPVIKDDVIYPQTVPVKEGDNVELPCDVTASPEAVVRWEMSQDDVIIPLDQRHVTDDQNTHRFTALWRDSGHYHCIAENALGTAKKTILVNVLVAPYIETPQSKTLTVRSGSTVKLACNVLYGNPAPSLKWKFINKDSTSRILRNNRTSSLTLNNVNKLNEGSYMCIVDNALGSDRIKYQLKVE
Summary
Description
Promotes cleavage furrow maturation during cytokinesis in preimplantation embryos. May play a role in the architecture of adhesive and flexible epithelial cell junctions. May play a role during myocardial remodeling by imparting an effect on cardiac fibroblast migration.
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Keywords
Age-related macular degeneration
Alternative splicing
Basement membrane
Calcium
Cell cycle
Cell division
Cell junction
Complete proteome
Cytoplasm
Disease mutation
Disulfide bond
EGF-like domain
Extracellular matrix
Glycoprotein
Immunoglobulin domain
Polymorphism
Reference proteome
Repeat
Secreted
Sensory transduction
Signal
Vision
Feature
chain Hemicentin-1
splice variant In isoform 3.
sequence variant In dbSNP:rs7539719.
splice variant In isoform 3.
sequence variant In dbSNP:rs7539719.
Uniprot
H9JIU1
A0A2H1VT69
A0A0N1PJI5
A0A0L7L9E3
A0A194PZJ6
A0A212F378
+ More
A0A2A4IXE1 A0A437BRF1 A0A437B1H8 A0A212F6F9 A0A0N1IPC9 A0A2A4J8M3 A0A0N1I874 A0A2H1V2M3 A0A3S2LTX9 A0A3S2L5P2 H9JIJ3 A0A194PYQ7 A0A158R5T7 A0A2W1BAW8 G3NWN1 A0A3B4WM49 A0A194Q7A1 A0A3B4UEA3 G3H9S6 A0A2D0T6P7 A0A2U9C6G6 A0A3N0YC56 A0A0S7H0Q2 A0A0S7GZK4 A0A3P8WNL8 A0A087Y670 A0A210QUZ9 A0A1I7ZUY9 A0A2B4RR01 A0A1I7ZTZ0 W4Y7I7 A0A3Q2CT39 A0A158QVI2 A0A3S2N4W0 A0A2G9Q1G3 A0A1D5PQF5 A0A3Q3AIK5 A0A1D5PWR7 A0A0R3T0Q4 A0A1D5NYH4 A0A1D5PY27 A0A1D5PMN5 A0A2I4B518 A0A3B4E5X1 A0A0S7LGL7 A0A3B4G3M3 A0A2A4JKA5 A0A3Q1B6M5 A0A401SMA0 G1T5K0 A0A0K0EUT2 A0A146YJH4 M3YKJ2 I3JUM9 A0A2Y9DEM0 A0A147AWT0 A0A068XX77 A0A3P9NWC9 E4XJ18 A0A146YJD2 A0A146YJL7 A0A0N4ZRH6 H3ADQ7 A0A3Q2V180 A0A3Q7PIR6 A0A2Y9JST8 A0A091GDU5 Q96RW7-2 Q96RW7 A0A226NXN6 A0A146YLE1 A0A1U7UYH7 A0A146YKT7 A0A158REM4 E0VC72 A0A1S3KK60 A0A1S3KKA0 A0A1W5AAI4 F6R9N9 A0A3M0K585 A0A0P7X0M9 M3X1M2 A0A3Q7T881 A0A194PFK9 A0A1I7RWJ4 A0A1A7ZB19 A0A1I7U000 A0A1I7U001 A0A1A8AP34 M3ZJI9 A0A158Q3H3 A0A3B4Z1V9 G7MF24
A0A2A4IXE1 A0A437BRF1 A0A437B1H8 A0A212F6F9 A0A0N1IPC9 A0A2A4J8M3 A0A0N1I874 A0A2H1V2M3 A0A3S2LTX9 A0A3S2L5P2 H9JIJ3 A0A194PYQ7 A0A158R5T7 A0A2W1BAW8 G3NWN1 A0A3B4WM49 A0A194Q7A1 A0A3B4UEA3 G3H9S6 A0A2D0T6P7 A0A2U9C6G6 A0A3N0YC56 A0A0S7H0Q2 A0A0S7GZK4 A0A3P8WNL8 A0A087Y670 A0A210QUZ9 A0A1I7ZUY9 A0A2B4RR01 A0A1I7ZTZ0 W4Y7I7 A0A3Q2CT39 A0A158QVI2 A0A3S2N4W0 A0A2G9Q1G3 A0A1D5PQF5 A0A3Q3AIK5 A0A1D5PWR7 A0A0R3T0Q4 A0A1D5NYH4 A0A1D5PY27 A0A1D5PMN5 A0A2I4B518 A0A3B4E5X1 A0A0S7LGL7 A0A3B4G3M3 A0A2A4JKA5 A0A3Q1B6M5 A0A401SMA0 G1T5K0 A0A0K0EUT2 A0A146YJH4 M3YKJ2 I3JUM9 A0A2Y9DEM0 A0A147AWT0 A0A068XX77 A0A3P9NWC9 E4XJ18 A0A146YJD2 A0A146YJL7 A0A0N4ZRH6 H3ADQ7 A0A3Q2V180 A0A3Q7PIR6 A0A2Y9JST8 A0A091GDU5 Q96RW7-2 Q96RW7 A0A226NXN6 A0A146YLE1 A0A1U7UYH7 A0A146YKT7 A0A158REM4 E0VC72 A0A1S3KK60 A0A1S3KKA0 A0A1W5AAI4 F6R9N9 A0A3M0K585 A0A0P7X0M9 M3X1M2 A0A3Q7T881 A0A194PFK9 A0A1I7RWJ4 A0A1A7ZB19 A0A1I7U000 A0A1I7U001 A0A1A8AP34 M3ZJI9 A0A158Q3H3 A0A3B4Z1V9 G7MF24
Pubmed
EMBL
BABH01023203
ODYU01004310
SOQ44045.1
KQ460426
KPJ14782.1
JTDY01002146
+ More
KOB72005.1 KQ459585 KPI98164.1 AGBW02010618 OWR48173.1 NWSH01005792 PCG63944.1 RSAL01000016 RVE53022.1 RSAL01000206 RVE44350.1 AGBW02010045 OWR49308.1 KPJ14781.1 NWSH01002649 PCG67762.1 KPJ14779.1 ODYU01000391 SOQ35095.1 RVE44349.1 RSAL01000133 RVE46324.1 BABH01027071 BABH01027072 BABH01027073 BABH01027074 BABH01027075 BABH01027076 BABH01027077 BABH01027078 BABH01027079 BABH01027080 KPI98163.1 KZ150210 PZC72208.1 KQ459337 KPJ01408.1 JH000237 EGV94970.1 CP026254 AWP10692.1 RJVU01048406 ROL43461.1 GBYX01448277 JAO33197.1 GBYX01448276 JAO33198.1 AYCK01004151 AYCK01004152 AYCK01004153 NEDP02001728 OWF52578.1 LSMT01000353 PFX19596.1 AAGJ04172486 AAGJ04172487 AAGJ04172488 AAGJ04172489 AAGJ04172490 UXSR01005435 VDD81984.1 CM012440 RVE73540.1 KV922518 PIO09001.1 AADN05000650 UZAE01000104 VDN96274.1 GBYX01170112 JAO88629.1 NWSH01001280 PCG71863.1 BEZZ01000368 GCC31496.1 AAGW02054358 AAGW02054359 AAGW02054360 AAGW02054361 AAGW02054362 AAGW02054363 GCES01032044 JAR54279.1 AEYP01068968 AEYP01068969 AEYP01068970 AEYP01068971 AEYP01068972 AEYP01068973 AEYP01068974 AEYP01068975 AEYP01068976 AEYP01068977 AERX01012991 AERX01012992 AERX01012993 AERX01012994 AERX01012995 GCES01003303 JAR83020.1 LN902843 CDS36997.1 FN653057 CBY10461.1 GCES01032043 JAR54280.1 GCES01032047 JAR54276.1 AFYH01199621 AFYH01199622 AFYH01199623 AFYH01199624 AFYH01199625 AFYH01199626 AFYH01199627 AFYH01199628 AFYH01199629 AFYH01199630 KL448046 KFO79531.1 AF156100 AK056336 AK056557 AL118512 AL121996 AL133515 AL133553 AL135796 AL135797 AL391824 BX928748 AJ306906 AWGT02000278 OXB72513.1 GCES01032046 JAR54277.1 GCES01032048 JAR54275.1 UYWX01020384 VDM31969.1 DS235048 EEB10978.1 QRBI01000117 RMC08359.1 JARO02003647 KPP70078.1 AANG04001043 KQ459604 KPI92161.1 HADY01001021 SBP39506.1 HADY01018309 SBP56794.1 UYYG01001157 VDN56791.1 CM001253 EHH15721.1
KOB72005.1 KQ459585 KPI98164.1 AGBW02010618 OWR48173.1 NWSH01005792 PCG63944.1 RSAL01000016 RVE53022.1 RSAL01000206 RVE44350.1 AGBW02010045 OWR49308.1 KPJ14781.1 NWSH01002649 PCG67762.1 KPJ14779.1 ODYU01000391 SOQ35095.1 RVE44349.1 RSAL01000133 RVE46324.1 BABH01027071 BABH01027072 BABH01027073 BABH01027074 BABH01027075 BABH01027076 BABH01027077 BABH01027078 BABH01027079 BABH01027080 KPI98163.1 KZ150210 PZC72208.1 KQ459337 KPJ01408.1 JH000237 EGV94970.1 CP026254 AWP10692.1 RJVU01048406 ROL43461.1 GBYX01448277 JAO33197.1 GBYX01448276 JAO33198.1 AYCK01004151 AYCK01004152 AYCK01004153 NEDP02001728 OWF52578.1 LSMT01000353 PFX19596.1 AAGJ04172486 AAGJ04172487 AAGJ04172488 AAGJ04172489 AAGJ04172490 UXSR01005435 VDD81984.1 CM012440 RVE73540.1 KV922518 PIO09001.1 AADN05000650 UZAE01000104 VDN96274.1 GBYX01170112 JAO88629.1 NWSH01001280 PCG71863.1 BEZZ01000368 GCC31496.1 AAGW02054358 AAGW02054359 AAGW02054360 AAGW02054361 AAGW02054362 AAGW02054363 GCES01032044 JAR54279.1 AEYP01068968 AEYP01068969 AEYP01068970 AEYP01068971 AEYP01068972 AEYP01068973 AEYP01068974 AEYP01068975 AEYP01068976 AEYP01068977 AERX01012991 AERX01012992 AERX01012993 AERX01012994 AERX01012995 GCES01003303 JAR83020.1 LN902843 CDS36997.1 FN653057 CBY10461.1 GCES01032043 JAR54280.1 GCES01032047 JAR54276.1 AFYH01199621 AFYH01199622 AFYH01199623 AFYH01199624 AFYH01199625 AFYH01199626 AFYH01199627 AFYH01199628 AFYH01199629 AFYH01199630 KL448046 KFO79531.1 AF156100 AK056336 AK056557 AL118512 AL121996 AL133515 AL133553 AL135796 AL135797 AL391824 BX928748 AJ306906 AWGT02000278 OXB72513.1 GCES01032046 JAR54277.1 GCES01032048 JAR54275.1 UYWX01020384 VDM31969.1 DS235048 EEB10978.1 QRBI01000117 RMC08359.1 JARO02003647 KPP70078.1 AANG04001043 KQ459604 KPI92161.1 HADY01001021 SBP39506.1 HADY01018309 SBP56794.1 UYYG01001157 VDN56791.1 CM001253 EHH15721.1
Proteomes
UP000005204
UP000053240
UP000037510
UP000053268
UP000007151
UP000218220
+ More
UP000283053 UP000046393 UP000007635 UP000261360 UP000261420 UP000001075 UP000221080 UP000246464 UP000265120 UP000028760 UP000242188 UP000095287 UP000225706 UP000007110 UP000265020 UP000046399 UP000267029 UP000000539 UP000046398 UP000278807 UP000192220 UP000261440 UP000261460 UP000257160 UP000287033 UP000001811 UP000035680 UP000000715 UP000005207 UP000248480 UP000017246 UP000242638 UP000038045 UP000008672 UP000264840 UP000286641 UP000248482 UP000053760 UP000005640 UP000198419 UP000189704 UP000046396 UP000274429 UP000009046 UP000087266 UP000192224 UP000002279 UP000269221 UP000034805 UP000011712 UP000286640 UP000095284 UP000095282 UP000002852 UP000038040 UP000274756 UP000261400
UP000283053 UP000046393 UP000007635 UP000261360 UP000261420 UP000001075 UP000221080 UP000246464 UP000265120 UP000028760 UP000242188 UP000095287 UP000225706 UP000007110 UP000265020 UP000046399 UP000267029 UP000000539 UP000046398 UP000278807 UP000192220 UP000261440 UP000261460 UP000257160 UP000287033 UP000001811 UP000035680 UP000000715 UP000005207 UP000248480 UP000017246 UP000242638 UP000038045 UP000008672 UP000264840 UP000286641 UP000248482 UP000053760 UP000005640 UP000198419 UP000189704 UP000046396 UP000274429 UP000009046 UP000087266 UP000192224 UP000002279 UP000269221 UP000034805 UP000011712 UP000286640 UP000095284 UP000095282 UP000002852 UP000038040 UP000274756 UP000261400
PRIDE
Pfam
Interpro
IPR013783
Ig-like_fold
+ More
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR036465 vWFA_dom_sf
IPR001841 Znf_RING
IPR001604 DNA/RNA_non-sp_Endonuclease
IPR011993 PH-like_dom_sf
IPR006020 PTB/PI_dom
IPR033930 FAM43A/B_PTB
IPR002035 VWF_A
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000884 TSP1_rpt
IPR036383 TSP1_rpt_sf
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR001881 EGF-like_Ca-bd_dom
IPR013151 Immunoglobulin
IPR013106 Ig_V-set
IPR026823 cEGF
IPR006605 G2_nidogen/fibulin_G2F
IPR009017 GFP
IPR006612 THAP_Znf
IPR000387 TYR_PHOSPHATASE_dom
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR000242 PTPase_domain
IPR029021 Prot-tyrosine_phosphatase-like
IPR003595 Tyr_Pase_cat
IPR016130 Tyr_Pase_AS
IPR032984 Hemicentin-2
IPR003006 Ig/MHC_CS
IPR013320 ConA-like_dom_sf
IPR002049 Laminin_EGF
IPR000034 Laminin_IV
IPR001791 Laminin_G
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR036465 vWFA_dom_sf
IPR001841 Znf_RING
IPR001604 DNA/RNA_non-sp_Endonuclease
IPR011993 PH-like_dom_sf
IPR006020 PTB/PI_dom
IPR033930 FAM43A/B_PTB
IPR002035 VWF_A
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000884 TSP1_rpt
IPR036383 TSP1_rpt_sf
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR001881 EGF-like_Ca-bd_dom
IPR013151 Immunoglobulin
IPR013106 Ig_V-set
IPR026823 cEGF
IPR006605 G2_nidogen/fibulin_G2F
IPR009017 GFP
IPR006612 THAP_Znf
IPR000387 TYR_PHOSPHATASE_dom
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR000242 PTPase_domain
IPR029021 Prot-tyrosine_phosphatase-like
IPR003595 Tyr_Pase_cat
IPR016130 Tyr_Pase_AS
IPR032984 Hemicentin-2
IPR003006 Ig/MHC_CS
IPR013320 ConA-like_dom_sf
IPR002049 Laminin_EGF
IPR000034 Laminin_IV
IPR001791 Laminin_G
SUPFAM
ProteinModelPortal
H9JIU1
A0A2H1VT69
A0A0N1PJI5
A0A0L7L9E3
A0A194PZJ6
A0A212F378
+ More
A0A2A4IXE1 A0A437BRF1 A0A437B1H8 A0A212F6F9 A0A0N1IPC9 A0A2A4J8M3 A0A0N1I874 A0A2H1V2M3 A0A3S2LTX9 A0A3S2L5P2 H9JIJ3 A0A194PYQ7 A0A158R5T7 A0A2W1BAW8 G3NWN1 A0A3B4WM49 A0A194Q7A1 A0A3B4UEA3 G3H9S6 A0A2D0T6P7 A0A2U9C6G6 A0A3N0YC56 A0A0S7H0Q2 A0A0S7GZK4 A0A3P8WNL8 A0A087Y670 A0A210QUZ9 A0A1I7ZUY9 A0A2B4RR01 A0A1I7ZTZ0 W4Y7I7 A0A3Q2CT39 A0A158QVI2 A0A3S2N4W0 A0A2G9Q1G3 A0A1D5PQF5 A0A3Q3AIK5 A0A1D5PWR7 A0A0R3T0Q4 A0A1D5NYH4 A0A1D5PY27 A0A1D5PMN5 A0A2I4B518 A0A3B4E5X1 A0A0S7LGL7 A0A3B4G3M3 A0A2A4JKA5 A0A3Q1B6M5 A0A401SMA0 G1T5K0 A0A0K0EUT2 A0A146YJH4 M3YKJ2 I3JUM9 A0A2Y9DEM0 A0A147AWT0 A0A068XX77 A0A3P9NWC9 E4XJ18 A0A146YJD2 A0A146YJL7 A0A0N4ZRH6 H3ADQ7 A0A3Q2V180 A0A3Q7PIR6 A0A2Y9JST8 A0A091GDU5 Q96RW7-2 Q96RW7 A0A226NXN6 A0A146YLE1 A0A1U7UYH7 A0A146YKT7 A0A158REM4 E0VC72 A0A1S3KK60 A0A1S3KKA0 A0A1W5AAI4 F6R9N9 A0A3M0K585 A0A0P7X0M9 M3X1M2 A0A3Q7T881 A0A194PFK9 A0A1I7RWJ4 A0A1A7ZB19 A0A1I7U000 A0A1I7U001 A0A1A8AP34 M3ZJI9 A0A158Q3H3 A0A3B4Z1V9 G7MF24
A0A2A4IXE1 A0A437BRF1 A0A437B1H8 A0A212F6F9 A0A0N1IPC9 A0A2A4J8M3 A0A0N1I874 A0A2H1V2M3 A0A3S2LTX9 A0A3S2L5P2 H9JIJ3 A0A194PYQ7 A0A158R5T7 A0A2W1BAW8 G3NWN1 A0A3B4WM49 A0A194Q7A1 A0A3B4UEA3 G3H9S6 A0A2D0T6P7 A0A2U9C6G6 A0A3N0YC56 A0A0S7H0Q2 A0A0S7GZK4 A0A3P8WNL8 A0A087Y670 A0A210QUZ9 A0A1I7ZUY9 A0A2B4RR01 A0A1I7ZTZ0 W4Y7I7 A0A3Q2CT39 A0A158QVI2 A0A3S2N4W0 A0A2G9Q1G3 A0A1D5PQF5 A0A3Q3AIK5 A0A1D5PWR7 A0A0R3T0Q4 A0A1D5NYH4 A0A1D5PY27 A0A1D5PMN5 A0A2I4B518 A0A3B4E5X1 A0A0S7LGL7 A0A3B4G3M3 A0A2A4JKA5 A0A3Q1B6M5 A0A401SMA0 G1T5K0 A0A0K0EUT2 A0A146YJH4 M3YKJ2 I3JUM9 A0A2Y9DEM0 A0A147AWT0 A0A068XX77 A0A3P9NWC9 E4XJ18 A0A146YJD2 A0A146YJL7 A0A0N4ZRH6 H3ADQ7 A0A3Q2V180 A0A3Q7PIR6 A0A2Y9JST8 A0A091GDU5 Q96RW7-2 Q96RW7 A0A226NXN6 A0A146YLE1 A0A1U7UYH7 A0A146YKT7 A0A158REM4 E0VC72 A0A1S3KK60 A0A1S3KKA0 A0A1W5AAI4 F6R9N9 A0A3M0K585 A0A0P7X0M9 M3X1M2 A0A3Q7T881 A0A194PFK9 A0A1I7RWJ4 A0A1A7ZB19 A0A1I7U000 A0A1I7U001 A0A1A8AP34 M3ZJI9 A0A158Q3H3 A0A3B4Z1V9 G7MF24
PDB
6IAA
E-value=6.50577e-29,
Score=319
Ontologies
GO
GO:0046872
GO:0016787
GO:0003676
GO:0005509
GO:0071711
GO:0033334
GO:0003677
GO:0016021
GO:0004725
GO:0005886
GO:0017134
GO:0005007
GO:0009617
GO:0030054
GO:0005938
GO:0005604
GO:0007049
GO:0051301
GO:0007156
GO:0038023
GO:0005201
GO:0007157
GO:0042803
GO:0062023
GO:0050839
GO:0005913
GO:0070062
GO:0007601
GO:0032154
GO:0050808
GO:0032589
GO:0005515
GO:0016705
GO:0020037
GO:0003917
GO:0016887
GO:0005634
GO:0030145
GO:0006412
GO:0016876
GO:0043039
PANTHER
Topology
Subcellular location
Secreted
The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Extracellular space The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Extracellular matrix The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Basement membrane The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cytoplasm The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cell junction The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cleavage furrow The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Extracellular space The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Extracellular matrix The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Basement membrane The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cytoplasm The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cell junction The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cleavage furrow The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
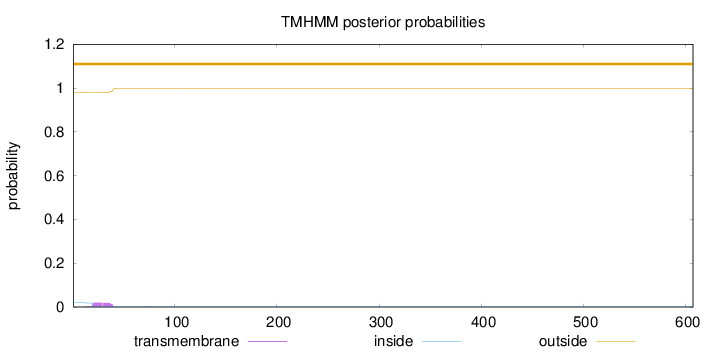
Length:
607
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.39696
Exp number, first 60 AAs:
0.39317
Total prob of N-in:
0.02070
outside
1 - 607
Population Genetic Test Statistics
Pi
208.256721
Theta
163.597795
Tajima's D
0.924658
CLR
0.236796
CSRT
0.642117894105295
Interpretation
Uncertain
