Gene
KWMTBOMO08260
Pre Gene Modal
BGIBMGA002329
Annotation
PREDICTED:_uncharacterized_protein_LOC103310341_[Acyrthosiphon_pisum]
Full name
ATP-dependent DNA helicase
Location in the cell
Extracellular Reliability : 1.296 Nuclear Reliability : 1.31
Sequence
CDS
ATGCCTAGAGGACGACGGGCGAACATCGGCCGCCGCACAAGACATGCAAGCCAGCAACAAGTGTATTCACAGAACTTAAGCGAAGAAAGACAAAATATAATAAGAGAAAATGCCCGATTGAGACAACGCGTGAGCACACGAAGATCATTGGCATCATACAATCGCTTGGCATTCCAATATGATCCCACTGCGAACTACAGTGATGATGAAAATTTAGATATTGGACCAATGACGACTATATGCCGATATTGCAATGCGTTAAAGTTCAAAAGAGAAACGGCTGGATTGTGCTGCGCAAGTGGAAAAGTCAAACTAGATCCATTACTTACACCACCACAGCCACTGAAACTATTGTTCGATGGAACTGATCCCGATTCCAGCCATTTTCTTCAACACATCCTTGAATACAATAACTGCTTTCGCATGACTTCCTTTGGCGCTAATATCATTCGAGAAGGCGGCTTCATGCCGACTTGCAAGCAATTCGCTGTCGACATGTATGTCAAAGTCGAGACTGAACGTTTAGCGTTCATCCGATTCAATCAGGCAAAGCTACGATCTGAGGACTATATACACTTGCGTGATGCTATTCATTCAGATGGTGATGTTCAAAATATTGGACGTCTGACGATTCTCCCATCATCTTATATCGGAAGCCCACGCCACATGCACGAATACGCTCAAGACGCTATGACGTACGTGCGAAATTATGGAACTCCGGATTTATTTATTACGTTCACATGCAATCCGAAGTGGACGGAAATTGAACCTGAAATTCCTGATCCAGTCACTGATCTCCATCTACACGATATTGTGACAACACAGATGGTGCATGGACCGTGCGGTGCATTAAATCCATTATCGCCTTGCATGGCTGATGGAAAGTGCACAAAACGATATCCGCGACCGTTAGTTGCTGAAACAGTCACAGGGAACGATGGATATCCAGTTTATCGTCGGCGTTCAAAAGAAAATAATGGTCGAACTATCAAAGTTAAAGTTCAAAATCAAGAGATTGAGATCGGAAATGAATTCATTGTACCATATTGCCCGCTGCTATCAAGAATTTTCGAAACACATGCAAACGTTGAGAGTTGTCATTCGGCCAAATCAATCAAATATTTGTGCAAGTACGTCACAAAAGGCAGCGACATGGCTGTGTTTGGTATTGCGTCGGAAAATGCGAATGACGAAATCAGTAACTTCCAAATGGACAGATACGTCAGTACTAATGAAGCACTGTGGCGATTATTGTCATTTCAAATTCATGAAAGATATCCCACAGTTGTACATTTAGCAGTGCATTTGGAAAATGGCCAAAGAGCTTACTTCACTGAGGCTAATGCGGCACAACGAGCTGAAAGACCACCATCGACAACATTGACTAGCTTCTTTGCAATGTGTGAAGCAGATCCATTCGCAGCGACGCTGATGTACGTTGAAATGCCCAAGTATTACACTTGGAATCAATCAACAAAGAAATTCCAACGTCGCAAACAAGGAACCCCAGTTCCAGATTGGCCACAGGTGTTTTCCACTGATGCACTAGGTCGCATGTATACTGTTCATCCTAGAAATGATGAATGTTTTTATTTGCGACTGCTGTTGGTAAATGTACGTGGACCAAAATCATTTGCGCATTTGAAAACTGTGAATGGCCACCAATGCCAAACATATCGAGAAGCATGTCAACTATTGGGTTTGCTGGAGAACGATTCTCATTGGGATTTAACACTTGCGGATTCAGTTGTTTCATCAAATGCGTACCAAATACGAACGCTGTTCGCAATTATCATCACCATATGTTTTCCTTCACAACCAATTCAGTAA
Protein
MPRGRRANIGRRTRHASQQQVYSQNLSEERQNIIRENARLRQRVSTRRSLASYNRLAFQYDPTANYSDDENLDIGPMTTICRYCNALKFKRETAGLCCASGKVKLDPLLTPPQPLKLLFDGTDPDSSHFLQHILEYNNCFRMTSFGANIIREGGFMPTCKQFAVDMYVKVETERLAFIRFNQAKLRSEDYIHLRDAIHSDGDVQNIGRLTILPSSYIGSPRHMHEYAQDAMTYVRNYGTPDLFITFTCNPKWTEIEPEIPDPVTDLHLHDIVTTQMVHGPCGALNPLSPCMADGKCTKRYPRPLVAETVTGNDGYPVYRRRSKENNGRTIKVKVQNQEIEIGNEFIVPYCPLLSRIFETHANVESCHSAKSIKYLCKYVTKGSDMAVFGIASENANDEISNFQMDRYVSTNEALWRLLSFQIHERYPTVVHLAVHLENGQRAYFTEANAAQRAERPPSTTLTSFFAMCEADPFAATLMYVEMPKYYTWNQSTKKFQRRKQGTPVPDWPQVFSTDALGRMYTVHPRNDECFYLRLLLVNVRGPKSFAHLKTVNGHQCQTYREACQLLGLLENDSHWDLTLADSVVSSNAYQIRTLFAIIITICFPSQPIQ
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
EC Number
3.6.4.12
EMBL
ABLF02017703
ABLF02041883
ABLF02039418
ABLF02041882
KN740416
KIH53763.1
+ More
KN761288 KIH48241.1 JOJR01000111 RCN45090.1 ABLF02018127 JOJR01000723 RCN34924.1 KN767628 KIH47497.1 LBMM01013912 KMQ85421.1 RSAL01000607 RVE41317.1 ABLF02024220 ABLF02041348 NIVC01001383 PAA68602.1 OIVN01003121 SPD08951.1 BEXD01002177 GBB97283.1 AGNK02004404 KZ305055 PIA34361.1 LR031875 VDD29540.1
KN761288 KIH48241.1 JOJR01000111 RCN45090.1 ABLF02018127 JOJR01000723 RCN34924.1 KN767628 KIH47497.1 LBMM01013912 KMQ85421.1 RSAL01000607 RVE41317.1 ABLF02024220 ABLF02041348 NIVC01001383 PAA68602.1 OIVN01003121 SPD08951.1 BEXD01002177 GBB97283.1 AGNK02004404 KZ305055 PIA34361.1 LR031875 VDD29540.1
Proteomes
Interpro
ProteinModelPortal
Ontologies
Topology
Subcellular location
Nucleus
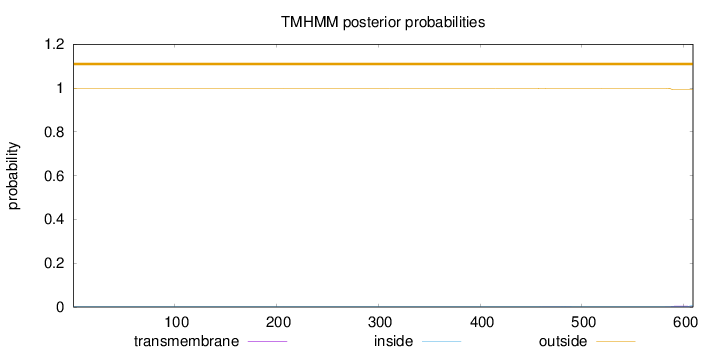
Length:
609
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11057
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00146
outside
1 - 609
Population Genetic Test Statistics
Pi
83.263275
Theta
53.005254
Tajima's D
2.155888
CLR
0.177595
CSRT
0.905304734763262
Interpretation
Uncertain
