Gene
KWMTBOMO08259
Pre Gene Modal
BGIBMGA009522
Annotation
PREDICTED:_probable_cytochrome_P450_301a1?_mitochondrial_[Amyelois_transitella]
Full name
Probable cytochrome P450 301a1, mitochondrial
Alternative Name
CYPCCCIA1
Location in the cell
Mitochondrial Reliability : 2.225
Sequence
CDS
ATGGGTCGCGCTTTACGCTCCTTCGCGGCCTACGCCAGACCTATACAGCTTCAGAGCACACGTAACTCATCGTCATGCCCATTCTCTAAGCGGCAACGTTCCCAAATAGCCCCGACCGCGGAGTTAAACGAGGAGATATTCGCGAACGCCCGTCCCTATTCGGAAGTTCCCGGCCCTAGACCTATCCCTATTTTGGGGAATACGTGGCGAATGGTCCCCGTTATAGGGCAGTTCGATATATCCGAGTTCGCGAAAGTGACCAAACAGTTTCTGGATACGTATGGCCGAATCGTTCGGCTCGGCGGTTTGATCGGAAGACCCGATCTTTTGTTCGTATATGATGCTGACGAGATCGAGAGGATGTATAGGAGGGAGGGACCGACCCCCTTCAGGCCTGCGATGCCGTGTCTCGTCAAATACAAATCGGAAGTCAGGAAGGATTTCTTTGGCGAGCTCCCGGGCGTTGTTGGCGTACACGGTGATCAATGGCGTAGATTTCGTTCCAAGGTTCAACGACCCATACTCCAGCCTCAAACGGTGAAGAAGTACGTGGCACCAATCGAGCTGGTCACGGAGGACTTCATTAAGTACATGGTAGACGCCCGAGACGAGAACGGGGACTTGCCTCATGAGTTCGACAACGACATTCACAGATGGTCTTTGGAATGTATCGGCCGCGTCGCCTTGGACGTCCGGCTGGGTTGTCTGTCGCCACAGTTGAATAGCAATTCAGAACCGCAACGCATCATCGATGCCGCAAAGTTTGCTTTACGAAACGTTGCAGTTTTAGAACTAAAGGCGCCTTATTGGAGATACATACCGACACCGCTTTGGAGTAAATACGTGAATAACATGAACTTCTTTGTTGAGATTTGCAGCCGTTATATCAATGAGGCGCTTGAAAGGCTGAAGACGAAGAAAGTGACGTCAGAGAACGATCTCTCGCTGCTCGAACGGGTTCTGAGGAGCGAAGGCGATCCAAAGATAGCCACCATAATGGCGTTAGACTTGATCCTTGTTGGGATTGATACGATTTCGATGGCGGTTTGCTCGATCCTGTACCAAGCAGCAACAAGATTGGAGCAACAGGATAAGATGGCGGAGGAGATCAGAAGGGTCCTCCCGGATCCGAGCAAGCCTCTGAGCTACTCGGACTTGGACAAGCTGCATTACACGAAAGCTTTCGTCAGAGAAGTTTTTAGAATGTATTCAACGGTCATCGGCAACGGTCGTACTCTGCAAGACGATGACGTCATATGTGGTTATCACATTCCAAAAGGTGTTCAAGTGGTGTTCCCGACGATCGTGACCGGCAACATGGAGCAGTTCGTCTCCGATCCTCTGGAGTTCAAGCCTGAGAGGTGGTTGGAAGGCGGCGGGAAGCTGCATCCATTCGCATCCCTGCCGTATGGCTTTGGCGCCCGGATATGTCTGGGACGGAGATTCGCTGACCTTGAGATCCAAGTGTTGTTGGCTAAGTTGCTGAGCCGTTACCGGTTGGAATACCACCACGAGCCCCTGGACTACGCCGTTACGTTCATGTACGCGCCGGACGGACCTCTGCGTCTGCGCATGATTGAACGATGA
Protein
MGRALRSFAAYARPIQLQSTRNSSSCPFSKRQRSQIAPTAELNEEIFANARPYSEVPGPRPIPILGNTWRMVPVIGQFDISEFAKVTKQFLDTYGRIVRLGGLIGRPDLLFVYDADEIERMYRREGPTPFRPAMPCLVKYKSEVRKDFFGELPGVVGVHGDQWRRFRSKVQRPILQPQTVKKYVAPIELVTEDFIKYMVDARDENGDLPHEFDNDIHRWSLECIGRVALDVRLGCLSPQLNSNSEPQRIIDAAKFALRNVAVLELKAPYWRYIPTPLWSKYVNNMNFFVEICSRYINEALERLKTKKVTSENDLSLLERVLRSEGDPKIATIMALDLILVGIDTISMAVCSILYQAATRLEQQDKMAEEIRRVLPDPSKPLSYSDLDKLHYTKAFVREVFRMYSTVIGNGRTLQDDDVICGYHIPKGVQVVFPTIVTGNMEQFVSDPLEFKPERWLEGGGKLHPFASLPYGFGARICLGRRFADLEIQVLLAKLLSRYRLEYHHEPLDYAVTFMYAPDGPLRLRMIER
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Keywords
Complete proteome
Heme
Iron
Membrane
Metal-binding
Mitochondrion
Monooxygenase
Oxidoreductase
Reference proteome
Transit peptide
Feature
chain Probable cytochrome P450 301a1, mitochondrial
Uniprot
L0N737
A0A2W1BQW2
A0A248QEJ0
A0A411E5A5
A0A0C5CGU0
A0A194PZJ1
+ More
A0A1V0D9E6 A0A286MXN0 A0A0N1PGZ2 A0A3S2ND10 A0A212FKZ2 X5D4J2 A0A411K711 A0A0N0BIJ6 A0A2A3EKE9 A0A088A413 A0A1W4XAA4 A0A2A3ELY0 E9IDI7 A0A1W6L1K6 K7J2R0 A0A0L7R620 A0A1I8M2B1 A0A2J7PYZ6 T1PI94 A0A3L8DGJ6 A0A026WY19 A0A0L0CMP0 A0A158NVG1 A0A151XBW6 A0A1W4VC89 B4KQT8 B4LL05 B4P582 A0A1B6H313 A0A3B0JNL2 B4HC64 B3MCT2 A0A1B0CHP8 W8BMR5 Q28YG9 A0A0A1WSD7 B4QDD3 B3NRY8 A0A195EC32 B4HPK9 A0A1I8QF68 Q9V6D6 A0A034W5D0 B4J9L1 A0A0M5JRA4 A0A0T6B368 A0A151I9G1 A0A232F1K2 A0A0M4E6Z7 D6QSF1 A0A1A9WNV8 A0A1B0BFL6 A0A1A9XAT3 B4NNE9 A0A1B6KMB4 A0A1A9V4S8 A0A1B0G1L5 E2AFK2 E0VHZ1 A0A2D1GSF3 A0A1S4G242 D2A102 A0A2M3Z1C2 A0A2M4BIB8 W5JQG0 B0WPH3 A0A023F3W6 A0A2M4BIG2 A0A2M4BJ81 N6TRD9 Q16FY4 A0A182FI14 A0A0K8VBZ7 A0A2M4A922 A0A2M4A8Q4 A0A182R7X5 A0A2S2R4W0 A0A182M7U6 A0A182NME1 A0A182Q9I3 A0A146M9Z8 J9JYT0 A0A2P8YRI1 A0A146LD79 A0A182WHR8 A0A182KP70 F4WNP8 Q7Q624 A0A182P0X9 A0A182XM70 A0A182IU57
A0A1V0D9E6 A0A286MXN0 A0A0N1PGZ2 A0A3S2ND10 A0A212FKZ2 X5D4J2 A0A411K711 A0A0N0BIJ6 A0A2A3EKE9 A0A088A413 A0A1W4XAA4 A0A2A3ELY0 E9IDI7 A0A1W6L1K6 K7J2R0 A0A0L7R620 A0A1I8M2B1 A0A2J7PYZ6 T1PI94 A0A3L8DGJ6 A0A026WY19 A0A0L0CMP0 A0A158NVG1 A0A151XBW6 A0A1W4VC89 B4KQT8 B4LL05 B4P582 A0A1B6H313 A0A3B0JNL2 B4HC64 B3MCT2 A0A1B0CHP8 W8BMR5 Q28YG9 A0A0A1WSD7 B4QDD3 B3NRY8 A0A195EC32 B4HPK9 A0A1I8QF68 Q9V6D6 A0A034W5D0 B4J9L1 A0A0M5JRA4 A0A0T6B368 A0A151I9G1 A0A232F1K2 A0A0M4E6Z7 D6QSF1 A0A1A9WNV8 A0A1B0BFL6 A0A1A9XAT3 B4NNE9 A0A1B6KMB4 A0A1A9V4S8 A0A1B0G1L5 E2AFK2 E0VHZ1 A0A2D1GSF3 A0A1S4G242 D2A102 A0A2M3Z1C2 A0A2M4BIB8 W5JQG0 B0WPH3 A0A023F3W6 A0A2M4BIG2 A0A2M4BJ81 N6TRD9 Q16FY4 A0A182FI14 A0A0K8VBZ7 A0A2M4A922 A0A2M4A8Q4 A0A182R7X5 A0A2S2R4W0 A0A182M7U6 A0A182NME1 A0A182Q9I3 A0A146M9Z8 J9JYT0 A0A2P8YRI1 A0A146LD79 A0A182WHR8 A0A182KP70 F4WNP8 Q7Q624 A0A182P0X9 A0A182XM70 A0A182IU57
EC Number
1.14.-.-
Pubmed
28756777
28881445
26354079
19954531
25299618
22118469
+ More
24361403 30716069 21282665 20075255 25315136 30249741 24508170 26108605 21347285 17994087 17550304 24495485 15632085 25830018 22936249 10731132 12537572 12537569 25348373 26319543 28648823 20798317 20566863 18362917 19820115 20920257 23761445 25474469 23537049 17510324 26823975 29403074 20966253 21719571 12364791 14747013 17210077
24361403 30716069 21282665 20075255 25315136 30249741 24508170 26108605 21347285 17994087 17550304 24495485 15632085 25830018 22936249 10731132 12537572 12537569 25348373 26319543 28648823 20798317 20566863 18362917 19820115 20920257 23761445 25474469 23537049 17510324 26823975 29403074 20966253 21719571 12364791 14747013 17210077
EMBL
AK289319
BAM73846.1
KZ149927
PZC77502.1
KX443474
ASO98049.1
+ More
MH236479 QBA57484.1 KP001112 AJN91157.1 KQ459585 KPI98159.1 KY212060 ARA91624.1 MF684348 ASX93982.1 KQ460426 KPJ14776.1 RSAL01000206 RVE44346.1 AGBW02007925 OWR54418.1 KF701181 AHW57351.1 MH500645 QBC73120.1 KQ435728 KOX77762.1 KZ288219 PBC32283.1 PBC32282.1 GL762454 EFZ21337.1 KX609532 ARN17939.1 KQ414648 KOC66320.1 NEVH01020341 PNF21558.1 KA648466 AFP63095.1 QOIP01000008 RLU19436.1 KK107085 EZA60039.1 JRES01000173 KNC33618.1 ADTU01002887 KQ982316 KYQ57839.1 CH933808 EDW09287.1 CH940648 EDW61812.2 CM000158 EDW90744.1 GECZ01000741 JAS69028.1 OUUW01000001 SPP74231.1 CH479278 EDW40113.1 CH902619 EDV37334.1 AJWK01012576 AJWK01012577 GAMC01011894 GAMC01011893 JAB94661.1 CM000071 EAL25996.1 GBXI01012984 JAD01308.1 CM000362 CM002911 EDX06798.1 KMY93249.1 CH954179 EDV56290.1 KQ979074 KYN22783.1 CH480816 EDW47593.1 AE013599 AY094897 GAKP01009627 JAC49325.1 CH916367 EDW02518.1 KR012855 ALD15930.1 LJIG01016032 KRT81820.1 KQ978277 KYM95701.1 NNAY01001258 OXU24595.1 CP012524 ALC42378.1 HM003575 ADF87561.1 JXJN01013540 CH964282 EDW85888.1 GEBQ01027394 JAT12583.1 CCAG010009655 GL439118 EFN67753.1 DS235172 EEB12997.1 MF471396 ATN96002.1 KQ971338 EFA02906.1 GGFM01001576 MBW22327.1 GGFJ01003641 MBW52782.1 ADMH02000632 ETN65528.1 DS232024 EDS32361.1 GBBI01002577 JAC16135.1 GGFJ01003642 MBW52783.1 GGFJ01003707 MBW52848.1 APGK01023061 APGK01023062 APGK01023063 APGK01023064 KB740462 ENN80608.1 CH478363 EAT33149.2 GDHF01015957 JAI36357.1 GGFK01003944 MBW37265.1 GGFK01003828 MBW37149.1 GGMS01015805 MBY85008.1 AXCM01009036 AXCN02000933 GDHC01002056 JAQ16573.1 ABLF02025279 PYGN01000410 PSN46858.1 GDHC01013597 JAQ05032.1 GL888237 EGI64241.1 AAAB01008960 EAA10792.4
MH236479 QBA57484.1 KP001112 AJN91157.1 KQ459585 KPI98159.1 KY212060 ARA91624.1 MF684348 ASX93982.1 KQ460426 KPJ14776.1 RSAL01000206 RVE44346.1 AGBW02007925 OWR54418.1 KF701181 AHW57351.1 MH500645 QBC73120.1 KQ435728 KOX77762.1 KZ288219 PBC32283.1 PBC32282.1 GL762454 EFZ21337.1 KX609532 ARN17939.1 KQ414648 KOC66320.1 NEVH01020341 PNF21558.1 KA648466 AFP63095.1 QOIP01000008 RLU19436.1 KK107085 EZA60039.1 JRES01000173 KNC33618.1 ADTU01002887 KQ982316 KYQ57839.1 CH933808 EDW09287.1 CH940648 EDW61812.2 CM000158 EDW90744.1 GECZ01000741 JAS69028.1 OUUW01000001 SPP74231.1 CH479278 EDW40113.1 CH902619 EDV37334.1 AJWK01012576 AJWK01012577 GAMC01011894 GAMC01011893 JAB94661.1 CM000071 EAL25996.1 GBXI01012984 JAD01308.1 CM000362 CM002911 EDX06798.1 KMY93249.1 CH954179 EDV56290.1 KQ979074 KYN22783.1 CH480816 EDW47593.1 AE013599 AY094897 GAKP01009627 JAC49325.1 CH916367 EDW02518.1 KR012855 ALD15930.1 LJIG01016032 KRT81820.1 KQ978277 KYM95701.1 NNAY01001258 OXU24595.1 CP012524 ALC42378.1 HM003575 ADF87561.1 JXJN01013540 CH964282 EDW85888.1 GEBQ01027394 JAT12583.1 CCAG010009655 GL439118 EFN67753.1 DS235172 EEB12997.1 MF471396 ATN96002.1 KQ971338 EFA02906.1 GGFM01001576 MBW22327.1 GGFJ01003641 MBW52782.1 ADMH02000632 ETN65528.1 DS232024 EDS32361.1 GBBI01002577 JAC16135.1 GGFJ01003642 MBW52783.1 GGFJ01003707 MBW52848.1 APGK01023061 APGK01023062 APGK01023063 APGK01023064 KB740462 ENN80608.1 CH478363 EAT33149.2 GDHF01015957 JAI36357.1 GGFK01003944 MBW37265.1 GGFK01003828 MBW37149.1 GGMS01015805 MBY85008.1 AXCM01009036 AXCN02000933 GDHC01002056 JAQ16573.1 ABLF02025279 PYGN01000410 PSN46858.1 GDHC01013597 JAQ05032.1 GL888237 EGI64241.1 AAAB01008960 EAA10792.4
Proteomes
UP000053268
UP000053240
UP000283053
UP000007151
UP000053105
UP000242457
+ More
UP000005203 UP000192223 UP000002358 UP000053825 UP000095301 UP000235965 UP000279307 UP000053097 UP000037069 UP000005205 UP000075809 UP000192221 UP000009192 UP000008792 UP000002282 UP000268350 UP000008744 UP000007801 UP000092461 UP000001819 UP000000304 UP000008711 UP000078492 UP000001292 UP000095300 UP000000803 UP000001070 UP000078542 UP000215335 UP000092553 UP000091820 UP000092460 UP000092443 UP000007798 UP000078200 UP000092444 UP000000311 UP000009046 UP000007266 UP000000673 UP000002320 UP000019118 UP000008820 UP000069272 UP000075900 UP000075883 UP000075884 UP000075886 UP000007819 UP000245037 UP000075920 UP000075882 UP000007755 UP000007062 UP000075885 UP000076407 UP000075880
UP000005203 UP000192223 UP000002358 UP000053825 UP000095301 UP000235965 UP000279307 UP000053097 UP000037069 UP000005205 UP000075809 UP000192221 UP000009192 UP000008792 UP000002282 UP000268350 UP000008744 UP000007801 UP000092461 UP000001819 UP000000304 UP000008711 UP000078492 UP000001292 UP000095300 UP000000803 UP000001070 UP000078542 UP000215335 UP000092553 UP000091820 UP000092460 UP000092443 UP000007798 UP000078200 UP000092444 UP000000311 UP000009046 UP000007266 UP000000673 UP000002320 UP000019118 UP000008820 UP000069272 UP000075900 UP000075883 UP000075884 UP000075886 UP000007819 UP000245037 UP000075920 UP000075882 UP000007755 UP000007062 UP000075885 UP000076407 UP000075880
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N737
A0A2W1BQW2
A0A248QEJ0
A0A411E5A5
A0A0C5CGU0
A0A194PZJ1
+ More
A0A1V0D9E6 A0A286MXN0 A0A0N1PGZ2 A0A3S2ND10 A0A212FKZ2 X5D4J2 A0A411K711 A0A0N0BIJ6 A0A2A3EKE9 A0A088A413 A0A1W4XAA4 A0A2A3ELY0 E9IDI7 A0A1W6L1K6 K7J2R0 A0A0L7R620 A0A1I8M2B1 A0A2J7PYZ6 T1PI94 A0A3L8DGJ6 A0A026WY19 A0A0L0CMP0 A0A158NVG1 A0A151XBW6 A0A1W4VC89 B4KQT8 B4LL05 B4P582 A0A1B6H313 A0A3B0JNL2 B4HC64 B3MCT2 A0A1B0CHP8 W8BMR5 Q28YG9 A0A0A1WSD7 B4QDD3 B3NRY8 A0A195EC32 B4HPK9 A0A1I8QF68 Q9V6D6 A0A034W5D0 B4J9L1 A0A0M5JRA4 A0A0T6B368 A0A151I9G1 A0A232F1K2 A0A0M4E6Z7 D6QSF1 A0A1A9WNV8 A0A1B0BFL6 A0A1A9XAT3 B4NNE9 A0A1B6KMB4 A0A1A9V4S8 A0A1B0G1L5 E2AFK2 E0VHZ1 A0A2D1GSF3 A0A1S4G242 D2A102 A0A2M3Z1C2 A0A2M4BIB8 W5JQG0 B0WPH3 A0A023F3W6 A0A2M4BIG2 A0A2M4BJ81 N6TRD9 Q16FY4 A0A182FI14 A0A0K8VBZ7 A0A2M4A922 A0A2M4A8Q4 A0A182R7X5 A0A2S2R4W0 A0A182M7U6 A0A182NME1 A0A182Q9I3 A0A146M9Z8 J9JYT0 A0A2P8YRI1 A0A146LD79 A0A182WHR8 A0A182KP70 F4WNP8 Q7Q624 A0A182P0X9 A0A182XM70 A0A182IU57
A0A1V0D9E6 A0A286MXN0 A0A0N1PGZ2 A0A3S2ND10 A0A212FKZ2 X5D4J2 A0A411K711 A0A0N0BIJ6 A0A2A3EKE9 A0A088A413 A0A1W4XAA4 A0A2A3ELY0 E9IDI7 A0A1W6L1K6 K7J2R0 A0A0L7R620 A0A1I8M2B1 A0A2J7PYZ6 T1PI94 A0A3L8DGJ6 A0A026WY19 A0A0L0CMP0 A0A158NVG1 A0A151XBW6 A0A1W4VC89 B4KQT8 B4LL05 B4P582 A0A1B6H313 A0A3B0JNL2 B4HC64 B3MCT2 A0A1B0CHP8 W8BMR5 Q28YG9 A0A0A1WSD7 B4QDD3 B3NRY8 A0A195EC32 B4HPK9 A0A1I8QF68 Q9V6D6 A0A034W5D0 B4J9L1 A0A0M5JRA4 A0A0T6B368 A0A151I9G1 A0A232F1K2 A0A0M4E6Z7 D6QSF1 A0A1A9WNV8 A0A1B0BFL6 A0A1A9XAT3 B4NNE9 A0A1B6KMB4 A0A1A9V4S8 A0A1B0G1L5 E2AFK2 E0VHZ1 A0A2D1GSF3 A0A1S4G242 D2A102 A0A2M3Z1C2 A0A2M4BIB8 W5JQG0 B0WPH3 A0A023F3W6 A0A2M4BIG2 A0A2M4BJ81 N6TRD9 Q16FY4 A0A182FI14 A0A0K8VBZ7 A0A2M4A922 A0A2M4A8Q4 A0A182R7X5 A0A2S2R4W0 A0A182M7U6 A0A182NME1 A0A182Q9I3 A0A146M9Z8 J9JYT0 A0A2P8YRI1 A0A146LD79 A0A182WHR8 A0A182KP70 F4WNP8 Q7Q624 A0A182P0X9 A0A182XM70 A0A182IU57
PDB
3K9Y
E-value=1.68399e-35,
Score=375
Ontologies
GO
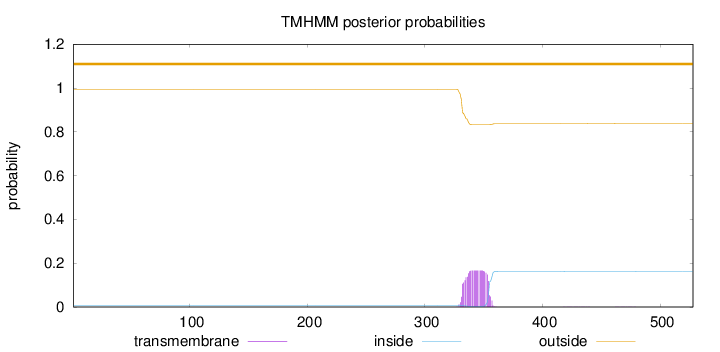
Topology
Subcellular location
Mitochondrion membrane
Length:
528
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.73977
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00673
outside
1 - 528
Population Genetic Test Statistics
Pi
20.79556
Theta
199.315244
Tajima's D
0.211475
CLR
1.11145
CSRT
0.428778561071946
Interpretation
Uncertain
