Gene
KWMTBOMO08257
Annotation
PREDICTED:_GDP-D-glucose_phosphorylase_1_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 2.083 Nuclear Reliability : 1.949
Sequence
CDS
ATGGAAAGACAAAATGATTTCAAAAATGTACCAAACTTATTCGAGGAAATACTTAAAAAAAAATGGGACGAAATACACGAGGGACCTAAAGTGTTTCGTTACAAAATTAAAGAAATAAATGAAAAATATTTACAAGAAAAATATCTAATTCAGTTAAATCCTGATCGTGGCAAAAAGAGAAGGGAGCCTGAAGCTATACACAGTCTTTCTCAACCGTTTGATGAAACAAAATTTAATTTTATGAAGGCATCATCTAATGAGATTTTACTAACTCTTATTAATGGAAATGAAGAACATGTTTTCTTGGTAAACGTAAGTCCAATTGCACGTTACCACATATTATTCTGCCCTTCCATGGGAAGTTGTCTTCCGCAAACAATAACACTAGATAGTCTAGAATTTGTTGCCGAAATGATGTTTGCATCGCAGGACCGTGATCTAAGAATTGGCTTCAACAGCCTCTGCGCATTTGCATCAGTCAATCATCTGCATTACCATTTATTATTTGAAAAGAGGACATTGTATATAGAGCAAGTGAAATGGAATCATTTAAAAGGACCTGTATATTTCAAGGAAGATTATCCTGTTCCAGCATTTTGTTGCAATATAAAAAAACTATCAGAAACTATCAATGCTTATATTCTTATAAAATATTTAGTGGATAAATCTATTGCTCACAATGTTTTAATAACAAATCGAAAATCACAAGAGACAGGAGTTCATGTTATAATTTGGCCTCGGAAAAAAACAGAAGGCGCCAAACAGTTTACAGATTTTAATGTTGCCATATGTGAACTCAGTGGATGGATTCCTATTTACGATGAAGAGACATACAGAAAAATAACAGCAGAACAAATTGAAGATGAATTAAAGAAATGGAAGTTAGATGATTTTTTTGAGCTATGTGAAGGAATCAAGTCCTTGTATTGA
Protein
MERQNDFKNVPNLFEEILKKKWDEIHEGPKVFRYKIKEINEKYLQEKYLIQLNPDRGKKRREPEAIHSLSQPFDETKFNFMKASSNEILLTLINGNEEHVFLVNVSPIARYHILFCPSMGSCLPQTITLDSLEFVAEMMFASQDRDLRIGFNSLCAFASVNHLHYHLLFEKRTLYIEQVKWNHLKGPVYFKEDYPVPAFCCNIKKLSETINAYILIKYLVDKSIAHNVLITNRKSQETGVHVIIWPRKKTEGAKQFTDFNVAICELSGWIPIYDEETYRKITAEQIEDELKKWKLDDFFELCEGIKSLY
Summary
Uniprot
A0A0N1PIB3
A0A2H1VC57
A0A2A4K8G0
A0A2W1BR17
A0A0N1I380
S4PK80
+ More
A0A212FL46 V4CHL4 A0A232EYD1 A0A2R5LC76 A0A2J7PZT0 A0A023GIW8 G3MPU4 K7JEN9 A0A0L7QPH6 A0A1Z5KX40 A0A1B6FSK5 A0A1E1XV02 A0A026WV65 A0A1B6JDW4 A0A023FKZ3 L7MM46 A0A3L8D9J4 A0A0K8TR75 A0A1B6I1Z6 A0A1B6E461 A0A2P8Y536 A0A210Q3S8 A0A1E1X625 A0A158NA10 A0A131Z6Q9 A0A154PDL4 T1JLX4 F4WC51 V5IF92 A0A151WH84 Q16Q87 A0A2L2Y6F5 A0A1S4FT07 A0A131XJQ4 A0A2H8TZZ3 A0A336LIL9 A0A369S7V7 B3RJU5 A0A195CUU9 A0A0L0C653 B3NCH0 A0A1A9WTA9 C4WWB8 A7SA65 A0A336LMW2 J9K070 B4N550 A0A1I8PXV9 R7TH55 B7QKM6 B4PEW0 A0A0J9RTM2 A0A023FYJ2 A0A0C9R9F1 A0A1B0FBG7 K7FA10 A0A1B6LSQ1 B4HL47 A0A444SDR4 A0A182GL93 B4H1Y0 Q8T4A7 A0A3Q3X5N0 Q9VSZ5 A0A0M4EY76 A0A195BK97 A0A2U9BZ17 A0A2T7NYM4 A0A2U9BWH0 Q29EF0 A0A226DBA0 A0A1B0BC44 A0A1W4U8R3 B0X8E0 A0A195ELG7 A0A1A9Y055 A0A2A3ENG5 A0A0K8TYC0 A0A2U9BX44 A0A401P1W8 A0A1J1I6C1 A0A1A9UG16 A0A1D2M4S6 A0A3S0ZWE9 A0A0V0GC68 A0A1B0DFM8 B4IXD4 A0A0P6FX87 A0A034VQD4 A0A0P5MGV6 G3WC22 A0A3Q3E0L3 A0A0N8CGB7
A0A212FL46 V4CHL4 A0A232EYD1 A0A2R5LC76 A0A2J7PZT0 A0A023GIW8 G3MPU4 K7JEN9 A0A0L7QPH6 A0A1Z5KX40 A0A1B6FSK5 A0A1E1XV02 A0A026WV65 A0A1B6JDW4 A0A023FKZ3 L7MM46 A0A3L8D9J4 A0A0K8TR75 A0A1B6I1Z6 A0A1B6E461 A0A2P8Y536 A0A210Q3S8 A0A1E1X625 A0A158NA10 A0A131Z6Q9 A0A154PDL4 T1JLX4 F4WC51 V5IF92 A0A151WH84 Q16Q87 A0A2L2Y6F5 A0A1S4FT07 A0A131XJQ4 A0A2H8TZZ3 A0A336LIL9 A0A369S7V7 B3RJU5 A0A195CUU9 A0A0L0C653 B3NCH0 A0A1A9WTA9 C4WWB8 A7SA65 A0A336LMW2 J9K070 B4N550 A0A1I8PXV9 R7TH55 B7QKM6 B4PEW0 A0A0J9RTM2 A0A023FYJ2 A0A0C9R9F1 A0A1B0FBG7 K7FA10 A0A1B6LSQ1 B4HL47 A0A444SDR4 A0A182GL93 B4H1Y0 Q8T4A7 A0A3Q3X5N0 Q9VSZ5 A0A0M4EY76 A0A195BK97 A0A2U9BZ17 A0A2T7NYM4 A0A2U9BWH0 Q29EF0 A0A226DBA0 A0A1B0BC44 A0A1W4U8R3 B0X8E0 A0A195ELG7 A0A1A9Y055 A0A2A3ENG5 A0A0K8TYC0 A0A2U9BX44 A0A401P1W8 A0A1J1I6C1 A0A1A9UG16 A0A1D2M4S6 A0A3S0ZWE9 A0A0V0GC68 A0A1B0DFM8 B4IXD4 A0A0P6FX87 A0A034VQD4 A0A0P5MGV6 G3WC22 A0A3Q3E0L3 A0A0N8CGB7
Pubmed
26354079
28756777
23622113
22118469
23254933
28648823
+ More
22216098 20075255 28528879 29209593 24508170 25576852 30249741 26369729 29403074 28812685 28503490 21347285 26830274 21719571 25765539 17510324 26561354 28049606 30042472 18719581 26108605 17994087 17615350 17550304 22936249 17381049 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 30297745 27289101 25348373 21709235
22216098 20075255 28528879 29209593 24508170 25576852 30249741 26369729 29403074 28812685 28503490 21347285 26830274 21719571 25765539 17510324 26561354 28049606 30042472 18719581 26108605 17994087 17615350 17550304 22936249 17381049 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 30297745 27289101 25348373 21709235
EMBL
KQ460426
KPJ14774.1
ODYU01001573
SOQ37982.1
NWSH01000071
PCG79960.1
+ More
KZ149927 PZC77539.1 KQ459256 KPJ02069.1 GAIX01004650 JAA87910.1 AGBW02007925 OWR54420.1 KB200454 ESP01625.1 NNAY01001637 OXU23353.1 GGLE01002976 MBY07102.1 NEVH01020332 PNF21845.1 GBBM01001511 JAC33907.1 JO843895 AEO35512.1 KQ414821 KOC60459.1 GFJQ02007609 GFJQ02000416 JAV99360.1 GECZ01016668 JAS53101.1 GFAA01000488 JAU02947.1 KK107105 EZA59551.1 GECU01010312 JAS97394.1 GBBK01002802 JAC21680.1 GACK01000725 JAA64309.1 QOIP01000011 RLU16578.1 GDAI01000975 JAI16628.1 GECU01026760 JAS80946.1 GEDC01004575 JAS32723.1 PYGN01000918 PSN39365.1 NEDP02005114 OWF43365.1 GFAC01004475 JAT94713.1 ADTU01009933 GEDV01001523 JAP87034.1 KQ434869 KZC09338.1 JH432212 GL888070 EGI68031.1 GANP01008842 JAB75626.1 KQ983135 KYQ47165.1 CH477757 EAT36557.1 IAAA01018561 IAAA01018562 LAA03718.1 GEFH01001999 JAP66582.1 GFXV01008042 MBW19847.1 UFQT01000014 SSX17696.1 NOWV01000045 RDD42988.1 DS985241 EDV29845.1 KQ977276 KYN04430.1 JRES01000850 KNC27736.1 CH954178 EDV51200.2 AK341972 BAH72188.1 DS469607 EDO39439.1 SSX17697.1 ABLF02037634 ABLF02037635 CH964101 EDW79489.1 AMQN01013027 KB309911 ELT93039.1 ABJB010178864 ABJB010298124 ABJB010680020 ABJB010685717 ABJB010876631 ABJB011025363 DS961247 EEC19398.1 CM000159 EDW93015.2 CM002912 KMY98659.1 GBBL01001507 JAC25813.1 GBYB01009542 JAG79309.1 CCAG010019566 AGCU01123460 GEBQ01013333 JAT26644.1 CH480815 EDW40866.1 SAUD01040768 RXG51546.1 JXUM01014237 KQ560397 KXJ82630.1 CH479203 EDW30332.1 AY089279 AAL90017.1 AE014296 AAF50263.1 ACL83281.1 CP012525 ALC43412.1 KQ976455 KYM85073.1 CP026252 AWP08679.1 PZQS01000008 PVD26233.1 AWP08678.1 CH379070 EAL30111.3 LNIX01000025 OXA42802.1 JXJN01011846 DS232485 EDS42476.1 KQ978739 KYN28724.1 KZ288206 PBC33044.1 GDHF01033224 GDHF01017262 JAI19090.1 JAI35052.1 AWP08680.1 AWP08681.1 BFAA01007078 GCB67139.1 CVRI01000041 CRK95274.1 LJIJ01004420 ODM87911.1 RQTK01000104 RUS87651.1 GECL01000417 JAP05707.1 AJVK01015130 CH916366 EDV96371.1 GDIQ01052433 JAN42304.1 GAKP01014640 JAC44312.1 GDIQ01257071 GDIQ01205906 GDIQ01204355 GDIQ01167569 GDIQ01044427 JAK84156.1 AEFK01006232 GDIP01134511 LRGB01001409 JAL69203.1 KZS12058.1
KZ149927 PZC77539.1 KQ459256 KPJ02069.1 GAIX01004650 JAA87910.1 AGBW02007925 OWR54420.1 KB200454 ESP01625.1 NNAY01001637 OXU23353.1 GGLE01002976 MBY07102.1 NEVH01020332 PNF21845.1 GBBM01001511 JAC33907.1 JO843895 AEO35512.1 KQ414821 KOC60459.1 GFJQ02007609 GFJQ02000416 JAV99360.1 GECZ01016668 JAS53101.1 GFAA01000488 JAU02947.1 KK107105 EZA59551.1 GECU01010312 JAS97394.1 GBBK01002802 JAC21680.1 GACK01000725 JAA64309.1 QOIP01000011 RLU16578.1 GDAI01000975 JAI16628.1 GECU01026760 JAS80946.1 GEDC01004575 JAS32723.1 PYGN01000918 PSN39365.1 NEDP02005114 OWF43365.1 GFAC01004475 JAT94713.1 ADTU01009933 GEDV01001523 JAP87034.1 KQ434869 KZC09338.1 JH432212 GL888070 EGI68031.1 GANP01008842 JAB75626.1 KQ983135 KYQ47165.1 CH477757 EAT36557.1 IAAA01018561 IAAA01018562 LAA03718.1 GEFH01001999 JAP66582.1 GFXV01008042 MBW19847.1 UFQT01000014 SSX17696.1 NOWV01000045 RDD42988.1 DS985241 EDV29845.1 KQ977276 KYN04430.1 JRES01000850 KNC27736.1 CH954178 EDV51200.2 AK341972 BAH72188.1 DS469607 EDO39439.1 SSX17697.1 ABLF02037634 ABLF02037635 CH964101 EDW79489.1 AMQN01013027 KB309911 ELT93039.1 ABJB010178864 ABJB010298124 ABJB010680020 ABJB010685717 ABJB010876631 ABJB011025363 DS961247 EEC19398.1 CM000159 EDW93015.2 CM002912 KMY98659.1 GBBL01001507 JAC25813.1 GBYB01009542 JAG79309.1 CCAG010019566 AGCU01123460 GEBQ01013333 JAT26644.1 CH480815 EDW40866.1 SAUD01040768 RXG51546.1 JXUM01014237 KQ560397 KXJ82630.1 CH479203 EDW30332.1 AY089279 AAL90017.1 AE014296 AAF50263.1 ACL83281.1 CP012525 ALC43412.1 KQ976455 KYM85073.1 CP026252 AWP08679.1 PZQS01000008 PVD26233.1 AWP08678.1 CH379070 EAL30111.3 LNIX01000025 OXA42802.1 JXJN01011846 DS232485 EDS42476.1 KQ978739 KYN28724.1 KZ288206 PBC33044.1 GDHF01033224 GDHF01017262 JAI19090.1 JAI35052.1 AWP08680.1 AWP08681.1 BFAA01007078 GCB67139.1 CVRI01000041 CRK95274.1 LJIJ01004420 ODM87911.1 RQTK01000104 RUS87651.1 GECL01000417 JAP05707.1 AJVK01015130 CH916366 EDV96371.1 GDIQ01052433 JAN42304.1 GAKP01014640 JAC44312.1 GDIQ01257071 GDIQ01205906 GDIQ01204355 GDIQ01167569 GDIQ01044427 JAK84156.1 AEFK01006232 GDIP01134511 LRGB01001409 JAL69203.1 KZS12058.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000007151
UP000030746
UP000215335
+ More
UP000235965 UP000002358 UP000053825 UP000053097 UP000279307 UP000245037 UP000242188 UP000005205 UP000076502 UP000007755 UP000075809 UP000008820 UP000253843 UP000009022 UP000078542 UP000037069 UP000008711 UP000091820 UP000001593 UP000007819 UP000007798 UP000095300 UP000014760 UP000001555 UP000002282 UP000092444 UP000007267 UP000001292 UP000288706 UP000069940 UP000249989 UP000008744 UP000261620 UP000000803 UP000092553 UP000078540 UP000246464 UP000245119 UP000001819 UP000198287 UP000092460 UP000192221 UP000002320 UP000078492 UP000092443 UP000242457 UP000288216 UP000183832 UP000078200 UP000094527 UP000271974 UP000092462 UP000001070 UP000007648 UP000261660 UP000076858
UP000235965 UP000002358 UP000053825 UP000053097 UP000279307 UP000245037 UP000242188 UP000005205 UP000076502 UP000007755 UP000075809 UP000008820 UP000253843 UP000009022 UP000078542 UP000037069 UP000008711 UP000091820 UP000001593 UP000007819 UP000007798 UP000095300 UP000014760 UP000001555 UP000002282 UP000092444 UP000007267 UP000001292 UP000288706 UP000069940 UP000249989 UP000008744 UP000261620 UP000000803 UP000092553 UP000078540 UP000246464 UP000245119 UP000001819 UP000198287 UP000092460 UP000192221 UP000002320 UP000078492 UP000092443 UP000242457 UP000288216 UP000183832 UP000078200 UP000094527 UP000271974 UP000092462 UP000001070 UP000007648 UP000261660 UP000076858
Pfam
Interpro
IPR036265
HIT-like_sf
+ More
IPR026506 GDPGP
IPR006578 MADF-dom
IPR012934 Znf_AD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR036034 PDZ_sf
IPR001478 PDZ
IPR019748 FERM_central
IPR029071 Ubiquitin-like_domsf
IPR000299 FERM_domain
IPR018980 FERM_PH-like_C
IPR019749 Band_41_domain
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR035963 FERM_2
IPR013969 Oligosacch_biosynth_Alg14
IPR019747 FERM_CS
IPR026506 GDPGP
IPR006578 MADF-dom
IPR012934 Znf_AD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR036034 PDZ_sf
IPR001478 PDZ
IPR019748 FERM_central
IPR029071 Ubiquitin-like_domsf
IPR000299 FERM_domain
IPR018980 FERM_PH-like_C
IPR019749 Band_41_domain
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR035963 FERM_2
IPR013969 Oligosacch_biosynth_Alg14
IPR019747 FERM_CS
SUPFAM
Gene 3D
ProteinModelPortal
A0A0N1PIB3
A0A2H1VC57
A0A2A4K8G0
A0A2W1BR17
A0A0N1I380
S4PK80
+ More
A0A212FL46 V4CHL4 A0A232EYD1 A0A2R5LC76 A0A2J7PZT0 A0A023GIW8 G3MPU4 K7JEN9 A0A0L7QPH6 A0A1Z5KX40 A0A1B6FSK5 A0A1E1XV02 A0A026WV65 A0A1B6JDW4 A0A023FKZ3 L7MM46 A0A3L8D9J4 A0A0K8TR75 A0A1B6I1Z6 A0A1B6E461 A0A2P8Y536 A0A210Q3S8 A0A1E1X625 A0A158NA10 A0A131Z6Q9 A0A154PDL4 T1JLX4 F4WC51 V5IF92 A0A151WH84 Q16Q87 A0A2L2Y6F5 A0A1S4FT07 A0A131XJQ4 A0A2H8TZZ3 A0A336LIL9 A0A369S7V7 B3RJU5 A0A195CUU9 A0A0L0C653 B3NCH0 A0A1A9WTA9 C4WWB8 A7SA65 A0A336LMW2 J9K070 B4N550 A0A1I8PXV9 R7TH55 B7QKM6 B4PEW0 A0A0J9RTM2 A0A023FYJ2 A0A0C9R9F1 A0A1B0FBG7 K7FA10 A0A1B6LSQ1 B4HL47 A0A444SDR4 A0A182GL93 B4H1Y0 Q8T4A7 A0A3Q3X5N0 Q9VSZ5 A0A0M4EY76 A0A195BK97 A0A2U9BZ17 A0A2T7NYM4 A0A2U9BWH0 Q29EF0 A0A226DBA0 A0A1B0BC44 A0A1W4U8R3 B0X8E0 A0A195ELG7 A0A1A9Y055 A0A2A3ENG5 A0A0K8TYC0 A0A2U9BX44 A0A401P1W8 A0A1J1I6C1 A0A1A9UG16 A0A1D2M4S6 A0A3S0ZWE9 A0A0V0GC68 A0A1B0DFM8 B4IXD4 A0A0P6FX87 A0A034VQD4 A0A0P5MGV6 G3WC22 A0A3Q3E0L3 A0A0N8CGB7
A0A212FL46 V4CHL4 A0A232EYD1 A0A2R5LC76 A0A2J7PZT0 A0A023GIW8 G3MPU4 K7JEN9 A0A0L7QPH6 A0A1Z5KX40 A0A1B6FSK5 A0A1E1XV02 A0A026WV65 A0A1B6JDW4 A0A023FKZ3 L7MM46 A0A3L8D9J4 A0A0K8TR75 A0A1B6I1Z6 A0A1B6E461 A0A2P8Y536 A0A210Q3S8 A0A1E1X625 A0A158NA10 A0A131Z6Q9 A0A154PDL4 T1JLX4 F4WC51 V5IF92 A0A151WH84 Q16Q87 A0A2L2Y6F5 A0A1S4FT07 A0A131XJQ4 A0A2H8TZZ3 A0A336LIL9 A0A369S7V7 B3RJU5 A0A195CUU9 A0A0L0C653 B3NCH0 A0A1A9WTA9 C4WWB8 A7SA65 A0A336LMW2 J9K070 B4N550 A0A1I8PXV9 R7TH55 B7QKM6 B4PEW0 A0A0J9RTM2 A0A023FYJ2 A0A0C9R9F1 A0A1B0FBG7 K7FA10 A0A1B6LSQ1 B4HL47 A0A444SDR4 A0A182GL93 B4H1Y0 Q8T4A7 A0A3Q3X5N0 Q9VSZ5 A0A0M4EY76 A0A195BK97 A0A2U9BZ17 A0A2T7NYM4 A0A2U9BWH0 Q29EF0 A0A226DBA0 A0A1B0BC44 A0A1W4U8R3 B0X8E0 A0A195ELG7 A0A1A9Y055 A0A2A3ENG5 A0A0K8TYC0 A0A2U9BX44 A0A401P1W8 A0A1J1I6C1 A0A1A9UG16 A0A1D2M4S6 A0A3S0ZWE9 A0A0V0GC68 A0A1B0DFM8 B4IXD4 A0A0P6FX87 A0A034VQD4 A0A0P5MGV6 G3WC22 A0A3Q3E0L3 A0A0N8CGB7
Ontologies
KEGG
GO
PANTHER
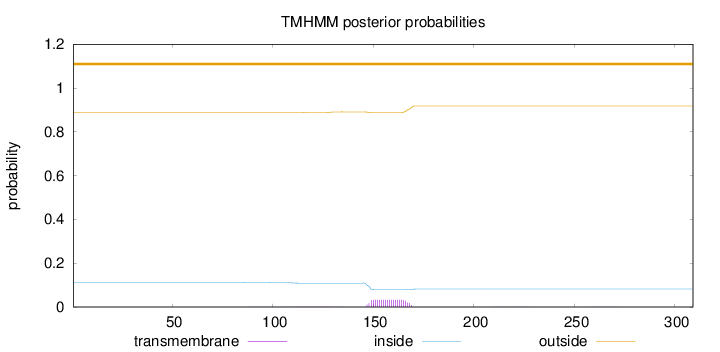
Topology
Length:
309
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.723600000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11135
outside
1 - 309
Population Genetic Test Statistics
Pi
153.282998
Theta
151.397362
Tajima's D
0.019262
CLR
89.761006
CSRT
0.381230938453077
Interpretation
Uncertain
