Gene
KWMTBOMO08254
Pre Gene Modal
BGIBMGA009520
Annotation
PREDICTED:_WD_repeat_domain_phosphoinositide-interacting_protein_4-like_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.88
Sequence
CDS
ATGAGGAGACGAGCTGGTGACGTGTCGAGCGTGCACCGCGACGGCGGCTCCAGCGCGCCCAGCGTGGTGCGCTGCCACCGCTCGGCGCTGGTGTGCGTGTGCGTCAGCGGCAGTGGCACCCGGCTCGCCACGGCGTCCGCGCGCGGCACCCTGGTCCGCCTCTGGGACGCCGTCTCCAGGCAGCTACTGCACGAGCTCAGGCGGGGCTCCGACTACGCTGACGTCTACTGTATAAGTCTGAGCGCTCAGGGCGGGCGCGCGGTGTGTGTGTCGGACAAGGGCACGGTGCACGTGTGGGCGCGCGGGACGCACGTGGCCGCCGCGCGGGCGCCGCCCGACACGCGCGCCCTCTGCGCCCTCACCGACGACAACAGCGCTGTCGTAGTGATATGCGAAGACGGCACCTTCCACAAGTTCACGTTTGCTGCGGAAGGAAACTGTCATCGGTCCGACTTTGAATATTTCTTGCAGGTTGGAGACGACGACGAATTCCTACAATGA
Protein
MRRRAGDVSSVHRDGGSSAPSVVRCHRSALVCVCVSGSGTRLATASARGTLVRLWDAVSRQLLHELRRGSDYADVYCISLSAQGGRAVCVSDKGTVHVWARGTHVAAARAPPDTRALCALTDDNSAVVVICEDGTFHKFTFAAEGNCHRSDFEYFLQVGDDDEFLQ
Summary
Uniprot
H9JJ19
A0A1E1WUC2
A0A2A4J4S2
A0A1E1W790
A0A437BPT6
A0A0N0PAJ5
+ More
A0A0N0PEF1 A0A212FE34 A0A2H1VAV9 A0A2W1BWF4 A0A0P5XI95 A0A0L7LLP2 A0A151WHQ4 A0A151IEN2 A0A195DML2 F4W6B1 A0A026VX33 A0A0P6HIE3 E2AVG5 A0A0N8ACD1 A0A0A9Z0H1 A0A0N8DG84 A0A195FIA6 A0A195AVP6 A0A158P229 E9FSU7 C3Z1P0 A0A1Y1LTI5 A0A0J7KF43 K7IYT4 E2BG11 A0A232EYI3 A0A2J7PFZ1 A0A1B6CCC6 A0A154PFZ4 A0A2A3E5L3 A0A088AFS1 A0A310SM71 A0A0P4VIS5 A0A0L7QQM6 A0A023F363 A0A069DS68 A0A0V0GEV0 A0A2P8Y0G2 A0A067RIE8 T1IAS7 A0A226E749 D6WAU4 E0VDQ6 A0A0P4W8H0 A0A1D2N9S0 A0A1B6K6H6 A0A1B6I1M0 W4ZAD8 A0A3R7QC68 A0A1B6KXL6 A0A1B6K8W7 A0A1B6ETR2 A0A0N0BGA0 C3Z7Q0 A0A2G8KM42 A0A2K6LPP0 A0A2S2Q9I6 A0A1D1W7K1 R7TLW8 A0A195EKD2 A0A195F677 A0A1U7S6F5 A7RXY8 A0A151X894 A0A3L8E5Q4 E2AYD7 F4WD07 A0A195BVW5 A0A158NKN7 H2XPX9 A0A1U7RZL5 A0A1S3J2E5 S4RER8 A0A2J7QN31 A0A154PPM5 E2BQR1 A0A0L8HH14 A0A151M7B9 A0A232FKR4 A0A2B4SU26 K7IZI5 A0A210PID3 A0A091CT11 V4BDR8 A0A3R7ST34 A0A3M6UY45 V9IH27 A0A088AEF1 A0A0L7QUQ4 A0A0L8HYR9 A0A2A3E648 A0A026WVE4 A0A087TFP7 A0A1E1XF07
A0A0N0PEF1 A0A212FE34 A0A2H1VAV9 A0A2W1BWF4 A0A0P5XI95 A0A0L7LLP2 A0A151WHQ4 A0A151IEN2 A0A195DML2 F4W6B1 A0A026VX33 A0A0P6HIE3 E2AVG5 A0A0N8ACD1 A0A0A9Z0H1 A0A0N8DG84 A0A195FIA6 A0A195AVP6 A0A158P229 E9FSU7 C3Z1P0 A0A1Y1LTI5 A0A0J7KF43 K7IYT4 E2BG11 A0A232EYI3 A0A2J7PFZ1 A0A1B6CCC6 A0A154PFZ4 A0A2A3E5L3 A0A088AFS1 A0A310SM71 A0A0P4VIS5 A0A0L7QQM6 A0A023F363 A0A069DS68 A0A0V0GEV0 A0A2P8Y0G2 A0A067RIE8 T1IAS7 A0A226E749 D6WAU4 E0VDQ6 A0A0P4W8H0 A0A1D2N9S0 A0A1B6K6H6 A0A1B6I1M0 W4ZAD8 A0A3R7QC68 A0A1B6KXL6 A0A1B6K8W7 A0A1B6ETR2 A0A0N0BGA0 C3Z7Q0 A0A2G8KM42 A0A2K6LPP0 A0A2S2Q9I6 A0A1D1W7K1 R7TLW8 A0A195EKD2 A0A195F677 A0A1U7S6F5 A7RXY8 A0A151X894 A0A3L8E5Q4 E2AYD7 F4WD07 A0A195BVW5 A0A158NKN7 H2XPX9 A0A1U7RZL5 A0A1S3J2E5 S4RER8 A0A2J7QN31 A0A154PPM5 E2BQR1 A0A0L8HH14 A0A151M7B9 A0A232FKR4 A0A2B4SU26 K7IZI5 A0A210PID3 A0A091CT11 V4BDR8 A0A3R7ST34 A0A3M6UY45 V9IH27 A0A088AEF1 A0A0L7QUQ4 A0A0L8HYR9 A0A2A3E648 A0A026WVE4 A0A087TFP7 A0A1E1XF07
Pubmed
19121390
26354079
22118469
28756777
26227816
21719571
+ More
24508170 20798317 25401762 21347285 21292972 18563158 28004739 20075255 28648823 27129103 25474469 26334808 29403074 24845553 18362917 19820115 20566863 27289101 29023486 27649274 23254933 17615350 30249741 12481130 15114417 22293439 28812685 30382153 28503490
24508170 20798317 25401762 21347285 21292972 18563158 28004739 20075255 28648823 27129103 25474469 26334808 29403074 24845553 18362917 19820115 20566863 27289101 29023486 27649274 23254933 17615350 30249741 12481130 15114417 22293439 28812685 30382153 28503490
EMBL
BABH01023169
GDQN01000628
JAT90426.1
NWSH01003080
PCG66981.1
GDQN01008239
+ More
JAT82815.1 RSAL01000021 RVE52416.1 KQ458751 KPJ05308.1 KQ459986 KPJ18840.1 AGBW02008987 OWR51968.1 ODYU01001573 SOQ37979.1 KZ149927 PZC77537.1 GDIP01071810 JAM31905.1 JTDY01000675 KOB76264.1 KQ983112 KYQ47347.1 KQ977858 KYM99304.1 KQ980724 KYN14072.1 GL887707 EGI70246.1 KK107670 EZA48195.1 GDIQ01032419 JAN62318.1 GL443111 EFN62575.1 GDIP01161289 LRGB01003123 JAJ62113.1 KZS04431.1 GBHO01008294 GBRD01000427 JAG35310.1 JAG65394.1 GDIP01036775 JAM66940.1 KQ981523 KYN40405.1 KQ976731 KYM76251.1 ADTU01006971 GL732524 EFX89757.1 GG666573 EEN53407.1 GEZM01048873 JAV76311.1 LBMM01008210 KMQ89053.1 GL448096 EFN85413.1 NNAY01001634 OXU23369.1 NEVH01025635 PNF15252.1 GEDC01026205 JAS11093.1 KQ434896 KZC10737.1 KZ288391 PBC26361.1 KQ763867 OAD54679.1 GDKW01002681 JAI53914.1 KQ414786 KOC60816.1 GBBI01003029 JAC15683.1 GBGD01002006 JAC86883.1 GECL01000322 JAP05802.1 PYGN01001092 PSN37664.1 KK852614 KDR20207.1 ACPB03005971 LNIX01000006 OXA52914.1 KQ971312 EEZ98685.1 DS235083 EEB11512.1 GDRN01081731 JAI61955.1 LJIJ01000130 ODN01998.1 GECU01000661 JAT07046.1 GECU01026883 JAS80823.1 AAGJ04041362 QCYY01003500 ROT63064.1 GEBQ01023822 JAT16155.1 GEBQ01032086 JAT07891.1 GECZ01028488 JAS41281.1 KQ435789 KOX74481.1 GG666591 EEN51405.1 MRZV01000482 PIK49082.1 GGMS01005206 MBY74409.1 BDGG01000022 GAV09355.1 AMQN01012122 KB309310 ELT94793.1 KQ978801 KYN28319.1 KQ981805 KYN35569.1 DS469551 EDO43705.1 KQ982431 KYQ56559.1 QOIP01000001 RLU27178.1 GL443887 EFN61554.1 GL888077 EGI67914.1 KQ976403 KYM92113.1 ADTU01002112 EAAA01001726 NEVH01013201 PNF29994.1 KQ435012 KZC13819.1 GL449798 EFN81980.1 KQ418170 KOF88532.1 AKHW03006417 KYO20422.1 NNAY01000090 OXU31089.1 LSMT01000020 PFX32649.1 NEDP02076656 OWF36241.1 KN124024 KFO22514.1 KB199905 ESP03877.1 QCYY01001985 ROT73816.1 RCHS01000482 RMX58616.1 JR047272 AEY60370.1 KQ414734 KOC62340.1 KQ416966 KOF94388.1 KZ288355 PBC27217.1 KK107087 EZA59997.1 KK115010 KFM63936.1 GFAC01001341 JAT97847.1
JAT82815.1 RSAL01000021 RVE52416.1 KQ458751 KPJ05308.1 KQ459986 KPJ18840.1 AGBW02008987 OWR51968.1 ODYU01001573 SOQ37979.1 KZ149927 PZC77537.1 GDIP01071810 JAM31905.1 JTDY01000675 KOB76264.1 KQ983112 KYQ47347.1 KQ977858 KYM99304.1 KQ980724 KYN14072.1 GL887707 EGI70246.1 KK107670 EZA48195.1 GDIQ01032419 JAN62318.1 GL443111 EFN62575.1 GDIP01161289 LRGB01003123 JAJ62113.1 KZS04431.1 GBHO01008294 GBRD01000427 JAG35310.1 JAG65394.1 GDIP01036775 JAM66940.1 KQ981523 KYN40405.1 KQ976731 KYM76251.1 ADTU01006971 GL732524 EFX89757.1 GG666573 EEN53407.1 GEZM01048873 JAV76311.1 LBMM01008210 KMQ89053.1 GL448096 EFN85413.1 NNAY01001634 OXU23369.1 NEVH01025635 PNF15252.1 GEDC01026205 JAS11093.1 KQ434896 KZC10737.1 KZ288391 PBC26361.1 KQ763867 OAD54679.1 GDKW01002681 JAI53914.1 KQ414786 KOC60816.1 GBBI01003029 JAC15683.1 GBGD01002006 JAC86883.1 GECL01000322 JAP05802.1 PYGN01001092 PSN37664.1 KK852614 KDR20207.1 ACPB03005971 LNIX01000006 OXA52914.1 KQ971312 EEZ98685.1 DS235083 EEB11512.1 GDRN01081731 JAI61955.1 LJIJ01000130 ODN01998.1 GECU01000661 JAT07046.1 GECU01026883 JAS80823.1 AAGJ04041362 QCYY01003500 ROT63064.1 GEBQ01023822 JAT16155.1 GEBQ01032086 JAT07891.1 GECZ01028488 JAS41281.1 KQ435789 KOX74481.1 GG666591 EEN51405.1 MRZV01000482 PIK49082.1 GGMS01005206 MBY74409.1 BDGG01000022 GAV09355.1 AMQN01012122 KB309310 ELT94793.1 KQ978801 KYN28319.1 KQ981805 KYN35569.1 DS469551 EDO43705.1 KQ982431 KYQ56559.1 QOIP01000001 RLU27178.1 GL443887 EFN61554.1 GL888077 EGI67914.1 KQ976403 KYM92113.1 ADTU01002112 EAAA01001726 NEVH01013201 PNF29994.1 KQ435012 KZC13819.1 GL449798 EFN81980.1 KQ418170 KOF88532.1 AKHW03006417 KYO20422.1 NNAY01000090 OXU31089.1 LSMT01000020 PFX32649.1 NEDP02076656 OWF36241.1 KN124024 KFO22514.1 KB199905 ESP03877.1 QCYY01001985 ROT73816.1 RCHS01000482 RMX58616.1 JR047272 AEY60370.1 KQ414734 KOC62340.1 KQ416966 KOF94388.1 KZ288355 PBC27217.1 KK107087 EZA59997.1 KK115010 KFM63936.1 GFAC01001341 JAT97847.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000075809 UP000078542 UP000078492 UP000007755 UP000053097 UP000000311 UP000076858 UP000078541 UP000078540 UP000005205 UP000000305 UP000001554 UP000036403 UP000002358 UP000008237 UP000215335 UP000235965 UP000076502 UP000242457 UP000005203 UP000053825 UP000245037 UP000027135 UP000015103 UP000198287 UP000007266 UP000009046 UP000094527 UP000007110 UP000283509 UP000053105 UP000230750 UP000233180 UP000186922 UP000014760 UP000189705 UP000001593 UP000279307 UP000008144 UP000085678 UP000245300 UP000053454 UP000050525 UP000225706 UP000242188 UP000028990 UP000030746 UP000275408 UP000054359
UP000037510 UP000075809 UP000078542 UP000078492 UP000007755 UP000053097 UP000000311 UP000076858 UP000078541 UP000078540 UP000005205 UP000000305 UP000001554 UP000036403 UP000002358 UP000008237 UP000215335 UP000235965 UP000076502 UP000242457 UP000005203 UP000053825 UP000245037 UP000027135 UP000015103 UP000198287 UP000007266 UP000009046 UP000094527 UP000007110 UP000283509 UP000053105 UP000230750 UP000233180 UP000186922 UP000014760 UP000189705 UP000001593 UP000279307 UP000008144 UP000085678 UP000245300 UP000053454 UP000050525 UP000225706 UP000242188 UP000028990 UP000030746 UP000275408 UP000054359
PRIDE
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JJ19
A0A1E1WUC2
A0A2A4J4S2
A0A1E1W790
A0A437BPT6
A0A0N0PAJ5
+ More
A0A0N0PEF1 A0A212FE34 A0A2H1VAV9 A0A2W1BWF4 A0A0P5XI95 A0A0L7LLP2 A0A151WHQ4 A0A151IEN2 A0A195DML2 F4W6B1 A0A026VX33 A0A0P6HIE3 E2AVG5 A0A0N8ACD1 A0A0A9Z0H1 A0A0N8DG84 A0A195FIA6 A0A195AVP6 A0A158P229 E9FSU7 C3Z1P0 A0A1Y1LTI5 A0A0J7KF43 K7IYT4 E2BG11 A0A232EYI3 A0A2J7PFZ1 A0A1B6CCC6 A0A154PFZ4 A0A2A3E5L3 A0A088AFS1 A0A310SM71 A0A0P4VIS5 A0A0L7QQM6 A0A023F363 A0A069DS68 A0A0V0GEV0 A0A2P8Y0G2 A0A067RIE8 T1IAS7 A0A226E749 D6WAU4 E0VDQ6 A0A0P4W8H0 A0A1D2N9S0 A0A1B6K6H6 A0A1B6I1M0 W4ZAD8 A0A3R7QC68 A0A1B6KXL6 A0A1B6K8W7 A0A1B6ETR2 A0A0N0BGA0 C3Z7Q0 A0A2G8KM42 A0A2K6LPP0 A0A2S2Q9I6 A0A1D1W7K1 R7TLW8 A0A195EKD2 A0A195F677 A0A1U7S6F5 A7RXY8 A0A151X894 A0A3L8E5Q4 E2AYD7 F4WD07 A0A195BVW5 A0A158NKN7 H2XPX9 A0A1U7RZL5 A0A1S3J2E5 S4RER8 A0A2J7QN31 A0A154PPM5 E2BQR1 A0A0L8HH14 A0A151M7B9 A0A232FKR4 A0A2B4SU26 K7IZI5 A0A210PID3 A0A091CT11 V4BDR8 A0A3R7ST34 A0A3M6UY45 V9IH27 A0A088AEF1 A0A0L7QUQ4 A0A0L8HYR9 A0A2A3E648 A0A026WVE4 A0A087TFP7 A0A1E1XF07
A0A0N0PEF1 A0A212FE34 A0A2H1VAV9 A0A2W1BWF4 A0A0P5XI95 A0A0L7LLP2 A0A151WHQ4 A0A151IEN2 A0A195DML2 F4W6B1 A0A026VX33 A0A0P6HIE3 E2AVG5 A0A0N8ACD1 A0A0A9Z0H1 A0A0N8DG84 A0A195FIA6 A0A195AVP6 A0A158P229 E9FSU7 C3Z1P0 A0A1Y1LTI5 A0A0J7KF43 K7IYT4 E2BG11 A0A232EYI3 A0A2J7PFZ1 A0A1B6CCC6 A0A154PFZ4 A0A2A3E5L3 A0A088AFS1 A0A310SM71 A0A0P4VIS5 A0A0L7QQM6 A0A023F363 A0A069DS68 A0A0V0GEV0 A0A2P8Y0G2 A0A067RIE8 T1IAS7 A0A226E749 D6WAU4 E0VDQ6 A0A0P4W8H0 A0A1D2N9S0 A0A1B6K6H6 A0A1B6I1M0 W4ZAD8 A0A3R7QC68 A0A1B6KXL6 A0A1B6K8W7 A0A1B6ETR2 A0A0N0BGA0 C3Z7Q0 A0A2G8KM42 A0A2K6LPP0 A0A2S2Q9I6 A0A1D1W7K1 R7TLW8 A0A195EKD2 A0A195F677 A0A1U7S6F5 A7RXY8 A0A151X894 A0A3L8E5Q4 E2AYD7 F4WD07 A0A195BVW5 A0A158NKN7 H2XPX9 A0A1U7RZL5 A0A1S3J2E5 S4RER8 A0A2J7QN31 A0A154PPM5 E2BQR1 A0A0L8HH14 A0A151M7B9 A0A232FKR4 A0A2B4SU26 K7IZI5 A0A210PID3 A0A091CT11 V4BDR8 A0A3R7ST34 A0A3M6UY45 V9IH27 A0A088AEF1 A0A0L7QUQ4 A0A0L8HYR9 A0A2A3E648 A0A026WVE4 A0A087TFP7 A0A1E1XF07
PDB
6IYY
E-value=1.08906e-25,
Score=284
Ontologies
GO
PANTHER
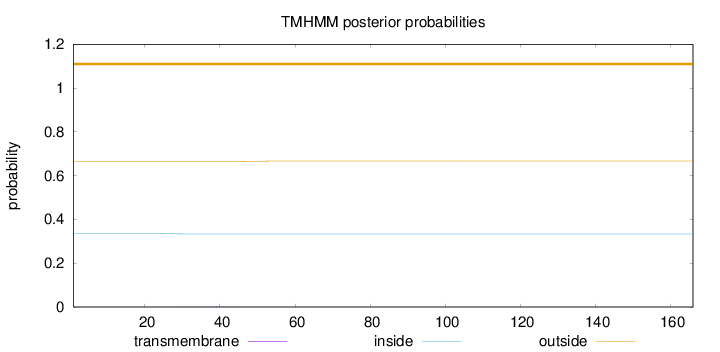
Topology
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0444300000000001
Exp number, first 60 AAs:
0.04389
Total prob of N-in:
0.33560
outside
1 - 166
Population Genetic Test Statistics
Pi
227.950479
Theta
171.129706
Tajima's D
0.884092
CLR
20.047531
CSRT
0.631868406579671
Interpretation
Uncertain
