Gene
KWMTBOMO08253
Pre Gene Modal
BGIBMGA009520
Annotation
PREDICTED:_WD_repeat_domain_phosphoinositide-interacting_protein_4-like_[Amyelois_transitella]
Full name
WD repeat domain phosphoinositide-interacting protein 4
Alternative Name
WD repeat-containing protein 45
WD repeat domain X-linked 1
WD repeat domain X-linked 1
Location in the cell
Nuclear Reliability : 3.161
Sequence
CDS
ATGGCGCGCCGGAAGAGCAGTGGCATAACGAGCTTGAGTTTCAATCAGGACCAAGGCTGTTTCACATGCTGTCTGAAGACTGGCCTGCGTGTCTATAATGTGGAGCCCCTGGTCGAGAAGGCGCATTACAGCAAAGAGGAGCTGGGCGAGGTGTCCCTATGCGAGATGGTGTTCCGCACCAACTGGCTGCTCGTGGTGAAAGCTCGGAAGCCGTACAGCCTCCTCCTGCTGGACGACCAGCAACGTGCCTTCAAGGCTGAGGTGGTCTTCAAGTCGCCGATACGAGCGTTGCGGGCCAGGAAGGATAAGGTGGCCGTTGTCCTGACGTCCAGCGTACAGGTGTTGGCGTTGCCGACGTTCCAGCGGGTCGCGTTGCTGCGGACGCCGTCCTCCGCGAGGCCGCTGTGCGCCATCGCCACGGAACCCAACGCGACGCAGATACTGGCCGCGCCCGCGCACCGCACCGGATCCCTGCAGTTGCTCGTATGTCCTAAACGAATTAGCTTGCCACTGAGAGAGAGAGAGAGAGAGGGAGAGAGAGTGACAGAGTATGCTTTATCTATATACTAG
Protein
MARRKSSGITSLSFNQDQGCFTCCLKTGLRVYNVEPLVEKAHYSKEELGEVSLCEMVFRTNWLLVVKARKPYSLLLLDDQQRAFKAEVVFKSPIRALRARKDKVAVVLTSSVQVLALPTFQRVALLRTPSSARPLCAIATEPNATQILAAPAHRTGSLQLLVCPKRISLPLREREREGERVTEYALSIY
Summary
Description
Component of the autophagy machinery that controls the major intracellular degradation process by which cytoplasmic materials are packaged into autophagosomes and delivered to lysosomes for degradation (PubMed:21802374). Activated by the STK11/AMPK signaling pathway upon starvation, WDR45 is involved in autophagosome assembly downstream of WIPI2, regulating the size of forming autophagosomes. Probably recruited to membranes through its PtdIns3P activity.
Component of the autophagy machinery that controls the major intracellular degradation process by which cytoplasmic materials are packaged into autophagosomes and delivered to lysosomes for degradation. Activated by the STK11/AMPK signaling pathway upon starvation, WDR45 is involved in autophagosome assembly downstream of WIPI2, regulating the size of forming autophagosomes. Probably recruited to membranes through its PtdIns3P activity.
Component of the autophagy machinery that controls the major intracellular degradation process by which cytoplasmic materials are packaged into autophagosomes and delivered to lysosomes for degradation. Activated by the STK11/AMPK signaling pathway upon starvation, WDR45 is involved in autophagosome assembly downstream of WIPI2, regulating the size of forming autophagosomes. Probably recruited to membranes through its PtdIns3P activity.
Subunit
Interacts with WIPI1. Interacts with WIPI2. Interacts with ATG2A and probably ATG2B. Interacts with ULK1. May interact with the PRKAA1, PRKAA2, PRKAB1 and PRKAG1 subunits of the AMPK kinase. May interact with NUDC.
Similarity
Belongs to the WD repeat SVP1 family.
Keywords
Autophagy
Complete proteome
Cytoplasm
Lipid-binding
Reference proteome
Repeat
WD repeat
Alternative splicing
Feature
chain WD repeat domain phosphoinositide-interacting protein 4
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JJ19
A0A2A4J4S2
A0A212FE34
A0A1E1W790
A0A437BPT6
A0A0N0PAJ5
+ More
A0A0N0PEF1 A0A2H1VAV9 A0A3L8DAL3 A0A026VX33 E2AVG5 A0A0J7KF43 A0A1B6ETR2 A0A232EYI3 D6WAU4 A0A195FIA6 E2BG11 A0A195DML2 A0A158P229 A0A1B6K8W7 V5FXJ8 A0A2A3E5L3 A0A195AVP6 F4W6B1 A0A1B6CCC6 A0A310SM71 A0A0N0BGA0 A0A088AFS1 A0A067RIE8 K7IYT4 A0A0L7QQM6 A0A2P8Y0G2 E9IV48 A0A1Y1LTI5 A0A087UBH3 E0VDQ6 A0A154PFZ4 A0A023F363 A0A1D2N9S0 A0A146LK63 A0A146M1P0 A0A0V0GEV0 A0A0A9X9H4 A0A0A9Z0H1 A0A226E749 T1IZY0 A0A0P4VIS5 T1IAS7 A0A069DS68 J3JU06 A0A0P5SKV8 A0A0P5T0P0 A0A1E1XQK1 E9FSU7 A0A061HZC9 A0A2J8R8E2 C9J5L0 Q5U2Y0 A0A1E1XFZ8 A0A2R5L6Z6 A0A061HU59 A0A140LJJ0 A0A3N0Z758 V9L4T8 G3MNZ3 A0A023GIP4 A0A0N8E9T0 A0A0P5DJI5 Q91VM3-2 H3D7R7 A0A0N8ACD1 A0A341DE06 A0A337SL11 A0A140TAA5 G3HNC1 A0A1U8CKM2 H3D7R2 A0A341DDB3 Q91VM3 A0A3Q4GJR9 A0A293M1X8 A0A0N8DKE1 V9KW62 A0A1A6HSC9 A0A131Y346 C1BLV3 G1U133 A0A3B4YFZ7 A0A2K6CD98 A0A2K6LMI5 A0A2K5V6B2 A0A2I2YM53 A0A2K6A9D5 A0A2R9CAA1 A0A2I3MNN2 A0A2K6PT20 A0A2K5KLI2
A0A0N0PEF1 A0A2H1VAV9 A0A3L8DAL3 A0A026VX33 E2AVG5 A0A0J7KF43 A0A1B6ETR2 A0A232EYI3 D6WAU4 A0A195FIA6 E2BG11 A0A195DML2 A0A158P229 A0A1B6K8W7 V5FXJ8 A0A2A3E5L3 A0A195AVP6 F4W6B1 A0A1B6CCC6 A0A310SM71 A0A0N0BGA0 A0A088AFS1 A0A067RIE8 K7IYT4 A0A0L7QQM6 A0A2P8Y0G2 E9IV48 A0A1Y1LTI5 A0A087UBH3 E0VDQ6 A0A154PFZ4 A0A023F363 A0A1D2N9S0 A0A146LK63 A0A146M1P0 A0A0V0GEV0 A0A0A9X9H4 A0A0A9Z0H1 A0A226E749 T1IZY0 A0A0P4VIS5 T1IAS7 A0A069DS68 J3JU06 A0A0P5SKV8 A0A0P5T0P0 A0A1E1XQK1 E9FSU7 A0A061HZC9 A0A2J8R8E2 C9J5L0 Q5U2Y0 A0A1E1XFZ8 A0A2R5L6Z6 A0A061HU59 A0A140LJJ0 A0A3N0Z758 V9L4T8 G3MNZ3 A0A023GIP4 A0A0N8E9T0 A0A0P5DJI5 Q91VM3-2 H3D7R7 A0A0N8ACD1 A0A341DE06 A0A337SL11 A0A140TAA5 G3HNC1 A0A1U8CKM2 H3D7R2 A0A341DDB3 Q91VM3 A0A3Q4GJR9 A0A293M1X8 A0A0N8DKE1 V9KW62 A0A1A6HSC9 A0A131Y346 C1BLV3 G1U133 A0A3B4YFZ7 A0A2K6CD98 A0A2K6LMI5 A0A2K5V6B2 A0A2I2YM53 A0A2K6A9D5 A0A2R9CAA1 A0A2I3MNN2 A0A2K6PT20 A0A2K5KLI2
Pubmed
19121390
22118469
26354079
30249741
24508170
20798317
+ More
28648823 18362917 19820115 21347285 21719571 24845553 20075255 29403074 21282665 28004739 20566863 25474469 27289101 26823975 25401762 27129103 26334808 22516182 23537049 29209593 21292972 23929341 15772651 15489334 21802374 28503490 19468303 21183079 24402279 22216098 10857745 16141072 15496914 17975172 15057822 21804562 29704459 25186727 21993624 22398555 22722832 25362486
28648823 18362917 19820115 21347285 21719571 24845553 20075255 29403074 21282665 28004739 20566863 25474469 27289101 26823975 25401762 27129103 26334808 22516182 23537049 29209593 21292972 23929341 15772651 15489334 21802374 28503490 19468303 21183079 24402279 22216098 10857745 16141072 15496914 17975172 15057822 21804562 29704459 25186727 21993624 22398555 22722832 25362486
EMBL
BABH01023169
NWSH01003080
PCG66981.1
AGBW02008987
OWR51968.1
GDQN01008239
+ More
JAT82815.1 RSAL01000021 RVE52416.1 KQ458751 KPJ05308.1 KQ459986 KPJ18840.1 ODYU01001573 SOQ37979.1 QOIP01000010 RLU17540.1 KK107670 EZA48195.1 GL443111 EFN62575.1 LBMM01008210 KMQ89053.1 GECZ01028488 JAS41281.1 NNAY01001634 OXU23369.1 KQ971312 EEZ98685.1 KQ981523 KYN40405.1 GL448096 EFN85413.1 KQ980724 KYN14072.1 ADTU01006971 GEBQ01032086 JAT07891.1 GALX01006076 JAB62390.1 KZ288391 PBC26361.1 KQ976731 KYM76251.1 GL887707 EGI70246.1 GEDC01026205 JAS11093.1 KQ763867 OAD54679.1 KQ435789 KOX74481.1 KK852614 KDR20207.1 KQ414786 KOC60816.1 PYGN01001092 PSN37664.1 GL766116 EFZ15578.1 GEZM01048873 JAV76311.1 KK119097 KFM74712.1 DS235083 EEB11512.1 KQ434896 KZC10737.1 GBBI01003029 JAC15683.1 LJIJ01000130 ODN01998.1 GDHC01011452 JAQ07177.1 GDHC01004901 JAQ13728.1 GECL01000322 JAP05802.1 GBHO01027308 JAG16296.1 GBHO01008294 GBRD01000427 JAG35310.1 JAG65394.1 LNIX01000006 OXA52914.1 JH431728 GDKW01002681 JAI53914.1 ACPB03005971 GBGD01002006 JAC86883.1 BT126717 KB632368 AEE61679.1 ERL93687.1 GDIP01138507 JAL65207.1 GDIP01137250 JAL66464.1 GFAA01001880 JAU01555.1 GL732524 EFX89757.1 KE685981 ERE63758.1 NDHI03003731 PNJ04776.1 AC231657 BC085816 GFAC01001179 JAT98009.1 GGLE01001119 MBY05245.1 ERE63759.1 AL671995 RJVU01007007 ROL54104.1 JW873627 AFP06144.1 JO843594 AEO35211.1 GBBM01001587 GBBM01001586 JAC33832.1 GDIQ01047828 JAN46909.1 GDIP01161290 GDIQ01118607 JAJ62112.1 JAL33119.1 AF229635 AK008427 AK145616 BC011479 GDIP01161289 LRGB01003123 JAJ62113.1 KZS04431.1 AANG04002273 AC130624 JH000543 RAZU01000328 EGW06093.1 RLQ59968.1 GFWV01008256 MAA32985.1 GDIP01025119 JAM78596.1 JW870280 AFP02798.1 LZPO01017290 OBS81124.1 GEFM01002868 JAP72928.1 BT075582 ACO10006.1 AAGW02040247 AAGW02040248 AAGW02040249 AQIA01084524 CABD030124443 CABD030124444 AJFE02020939 AHZZ02037104
JAT82815.1 RSAL01000021 RVE52416.1 KQ458751 KPJ05308.1 KQ459986 KPJ18840.1 ODYU01001573 SOQ37979.1 QOIP01000010 RLU17540.1 KK107670 EZA48195.1 GL443111 EFN62575.1 LBMM01008210 KMQ89053.1 GECZ01028488 JAS41281.1 NNAY01001634 OXU23369.1 KQ971312 EEZ98685.1 KQ981523 KYN40405.1 GL448096 EFN85413.1 KQ980724 KYN14072.1 ADTU01006971 GEBQ01032086 JAT07891.1 GALX01006076 JAB62390.1 KZ288391 PBC26361.1 KQ976731 KYM76251.1 GL887707 EGI70246.1 GEDC01026205 JAS11093.1 KQ763867 OAD54679.1 KQ435789 KOX74481.1 KK852614 KDR20207.1 KQ414786 KOC60816.1 PYGN01001092 PSN37664.1 GL766116 EFZ15578.1 GEZM01048873 JAV76311.1 KK119097 KFM74712.1 DS235083 EEB11512.1 KQ434896 KZC10737.1 GBBI01003029 JAC15683.1 LJIJ01000130 ODN01998.1 GDHC01011452 JAQ07177.1 GDHC01004901 JAQ13728.1 GECL01000322 JAP05802.1 GBHO01027308 JAG16296.1 GBHO01008294 GBRD01000427 JAG35310.1 JAG65394.1 LNIX01000006 OXA52914.1 JH431728 GDKW01002681 JAI53914.1 ACPB03005971 GBGD01002006 JAC86883.1 BT126717 KB632368 AEE61679.1 ERL93687.1 GDIP01138507 JAL65207.1 GDIP01137250 JAL66464.1 GFAA01001880 JAU01555.1 GL732524 EFX89757.1 KE685981 ERE63758.1 NDHI03003731 PNJ04776.1 AC231657 BC085816 GFAC01001179 JAT98009.1 GGLE01001119 MBY05245.1 ERE63759.1 AL671995 RJVU01007007 ROL54104.1 JW873627 AFP06144.1 JO843594 AEO35211.1 GBBM01001587 GBBM01001586 JAC33832.1 GDIQ01047828 JAN46909.1 GDIP01161290 GDIQ01118607 JAJ62112.1 JAL33119.1 AF229635 AK008427 AK145616 BC011479 GDIP01161289 LRGB01003123 JAJ62113.1 KZS04431.1 AANG04002273 AC130624 JH000543 RAZU01000328 EGW06093.1 RLQ59968.1 GFWV01008256 MAA32985.1 GDIP01025119 JAM78596.1 JW870280 AFP02798.1 LZPO01017290 OBS81124.1 GEFM01002868 JAP72928.1 BT075582 ACO10006.1 AAGW02040247 AAGW02040248 AAGW02040249 AQIA01084524 CABD030124443 CABD030124444 AJFE02020939 AHZZ02037104
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000279307 UP000053097 UP000000311 UP000036403 UP000215335 UP000007266 UP000078541 UP000008237 UP000078492 UP000005205 UP000242457 UP000078540 UP000007755 UP000053105 UP000005203 UP000027135 UP000002358 UP000053825 UP000245037 UP000054359 UP000009046 UP000076502 UP000094527 UP000198287 UP000015103 UP000030742 UP000000305 UP000030759 UP000005640 UP000002494 UP000000589 UP000007303 UP000076858 UP000252040 UP000011712 UP000001075 UP000273346 UP000189706 UP000261580 UP000092124 UP000001811 UP000261360 UP000233120 UP000233180 UP000233100 UP000001519 UP000233140 UP000240080 UP000028761 UP000233200 UP000233060
UP000279307 UP000053097 UP000000311 UP000036403 UP000215335 UP000007266 UP000078541 UP000008237 UP000078492 UP000005205 UP000242457 UP000078540 UP000007755 UP000053105 UP000005203 UP000027135 UP000002358 UP000053825 UP000245037 UP000054359 UP000009046 UP000076502 UP000094527 UP000198287 UP000015103 UP000030742 UP000000305 UP000030759 UP000005640 UP000002494 UP000000589 UP000007303 UP000076858 UP000252040 UP000011712 UP000001075 UP000273346 UP000189706 UP000261580 UP000092124 UP000001811 UP000261360 UP000233120 UP000233180 UP000233100 UP000001519 UP000233140 UP000240080 UP000028761 UP000233200 UP000233060
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JJ19
A0A2A4J4S2
A0A212FE34
A0A1E1W790
A0A437BPT6
A0A0N0PAJ5
+ More
A0A0N0PEF1 A0A2H1VAV9 A0A3L8DAL3 A0A026VX33 E2AVG5 A0A0J7KF43 A0A1B6ETR2 A0A232EYI3 D6WAU4 A0A195FIA6 E2BG11 A0A195DML2 A0A158P229 A0A1B6K8W7 V5FXJ8 A0A2A3E5L3 A0A195AVP6 F4W6B1 A0A1B6CCC6 A0A310SM71 A0A0N0BGA0 A0A088AFS1 A0A067RIE8 K7IYT4 A0A0L7QQM6 A0A2P8Y0G2 E9IV48 A0A1Y1LTI5 A0A087UBH3 E0VDQ6 A0A154PFZ4 A0A023F363 A0A1D2N9S0 A0A146LK63 A0A146M1P0 A0A0V0GEV0 A0A0A9X9H4 A0A0A9Z0H1 A0A226E749 T1IZY0 A0A0P4VIS5 T1IAS7 A0A069DS68 J3JU06 A0A0P5SKV8 A0A0P5T0P0 A0A1E1XQK1 E9FSU7 A0A061HZC9 A0A2J8R8E2 C9J5L0 Q5U2Y0 A0A1E1XFZ8 A0A2R5L6Z6 A0A061HU59 A0A140LJJ0 A0A3N0Z758 V9L4T8 G3MNZ3 A0A023GIP4 A0A0N8E9T0 A0A0P5DJI5 Q91VM3-2 H3D7R7 A0A0N8ACD1 A0A341DE06 A0A337SL11 A0A140TAA5 G3HNC1 A0A1U8CKM2 H3D7R2 A0A341DDB3 Q91VM3 A0A3Q4GJR9 A0A293M1X8 A0A0N8DKE1 V9KW62 A0A1A6HSC9 A0A131Y346 C1BLV3 G1U133 A0A3B4YFZ7 A0A2K6CD98 A0A2K6LMI5 A0A2K5V6B2 A0A2I2YM53 A0A2K6A9D5 A0A2R9CAA1 A0A2I3MNN2 A0A2K6PT20 A0A2K5KLI2
A0A0N0PEF1 A0A2H1VAV9 A0A3L8DAL3 A0A026VX33 E2AVG5 A0A0J7KF43 A0A1B6ETR2 A0A232EYI3 D6WAU4 A0A195FIA6 E2BG11 A0A195DML2 A0A158P229 A0A1B6K8W7 V5FXJ8 A0A2A3E5L3 A0A195AVP6 F4W6B1 A0A1B6CCC6 A0A310SM71 A0A0N0BGA0 A0A088AFS1 A0A067RIE8 K7IYT4 A0A0L7QQM6 A0A2P8Y0G2 E9IV48 A0A1Y1LTI5 A0A087UBH3 E0VDQ6 A0A154PFZ4 A0A023F363 A0A1D2N9S0 A0A146LK63 A0A146M1P0 A0A0V0GEV0 A0A0A9X9H4 A0A0A9Z0H1 A0A226E749 T1IZY0 A0A0P4VIS5 T1IAS7 A0A069DS68 J3JU06 A0A0P5SKV8 A0A0P5T0P0 A0A1E1XQK1 E9FSU7 A0A061HZC9 A0A2J8R8E2 C9J5L0 Q5U2Y0 A0A1E1XFZ8 A0A2R5L6Z6 A0A061HU59 A0A140LJJ0 A0A3N0Z758 V9L4T8 G3MNZ3 A0A023GIP4 A0A0N8E9T0 A0A0P5DJI5 Q91VM3-2 H3D7R7 A0A0N8ACD1 A0A341DE06 A0A337SL11 A0A140TAA5 G3HNC1 A0A1U8CKM2 H3D7R2 A0A341DDB3 Q91VM3 A0A3Q4GJR9 A0A293M1X8 A0A0N8DKE1 V9KW62 A0A1A6HSC9 A0A131Y346 C1BLV3 G1U133 A0A3B4YFZ7 A0A2K6CD98 A0A2K6LMI5 A0A2K5V6B2 A0A2I2YM53 A0A2K6A9D5 A0A2R9CAA1 A0A2I3MNN2 A0A2K6PT20 A0A2K5KLI2
PDB
6IYY
E-value=2.63116e-19,
Score=230
Ontologies
GO
PANTHER
Topology
Subcellular location
Preautophagosomal structure
Diffusely localized in the cytoplasm under nutrient-rich conditions. Localizes to autophagic structures during starvation-induced autophagy. With evidence from 1 publications.
Cytoplasm Diffusely localized in the cytoplasm under nutrient-rich conditions. Localizes to autophagic structures during starvation-induced autophagy. With evidence from 1 publications.
Cytoplasm Diffusely localized in the cytoplasm under nutrient-rich conditions. Localizes to autophagic structures during starvation-induced autophagy. With evidence from 1 publications.
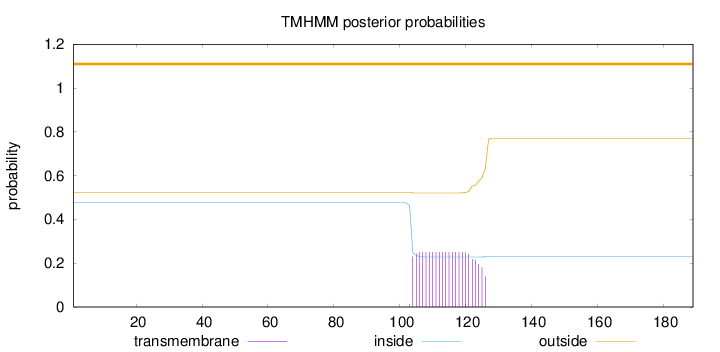
Length:
189
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.4463
Exp number, first 60 AAs:
0.00075
Total prob of N-in:
0.47636
outside
1 - 189
Population Genetic Test Statistics
Pi
257.396242
Theta
163.695794
Tajima's D
2.465917
CLR
0.239809
CSRT
0.941702914854257
Interpretation
Uncertain
