Gene
KWMTBOMO08251
Annotation
Yokozuna_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.018 Nuclear Reliability : 1.354
Sequence
CDS
ATGACAGCCGAGATGCTTCACACAGACATAGCAGGACCGATTAGACATCAAGGACCTAATGGTGAAAAATATTTTATGACAATTATTGACGATTTCTCTCATTTTTGCGTAGTGTATCCCCTGAAGAGCAAATCTGAAGCAACAGAACGACTGATGAACTATGTATTAAGAATTGAAAACGAAACGGGGAATAAAGTTAAAAGAATTAGATGCGATAATGGTGGAGAGTACACTGCTAATAGTTTGAAACTTTTTTGCATAAATAGAGGTATTAGTTTAGAATACACATTACCGTACACGCCACAACTTAATGGCATATCGGAACGGATGAACCGTACATTATGCGACAAAACAAGGACTCTATTAGCGGAAACCAATTTGCCAAAACATCTGTGGTGCGAAGCGATACAATGTGCAGTTTACCAATTAAACAGATCGCCATCATGGGCCATAAACTTCAAGACACCCAGCGGAATATTTTTTGGTAAAAACGACCTATCGAGATTGAGAGTATTTGGTTCCAAAGCTTGGGCTACTATAGTACCCAAACCAGACAAGTTAGCCAAGAGAGCAAAAGAAACAAGAATGGTAGGTTATGGAACTGTTGGTTACAGATTATGGGATCCAGAAGAAGATAAGATAGTTATTTCCAGAGATGTAAGATTTGATGAAAGCGACATTAAATTCAAAAATAACGAAGAAGTGAAACAAGTAAGATATGTTCAAGACAAAGAAGAAAAAATCAATAAAGAAAAGGAAGATATTGTAGAACAGGAAGATGAATTTAAAGATGCAGAAATGAACACTGATCAAATTGAAGAAGCAATAATCAGACCCAAAAGAACGATAAATCCTCCTGCTTATCTTAAAGATTACGAGACGAATATAGAAAACATAAATGAAACTAATATTGTTATATGTCTGAGTGCAAATGCACCAGAAACTTTCGAAGAAGCTATAAAAGAACCAGAATGGAAAAACGCAATCAATAAAGAGCTAGATGCACTCCAAAGACTACAAACGTGGGAAGAACAAGAAATAACCGATGGAAGAAAGCTCATAGATACAAAATGGATATTTAAACAAAAAGAAGATGGAACAAAAAAGGCAAGACTGGTAGCTAGAGGCTTCCAACAAAAGAAAGAAGATATTTTTGACAGCGTTTATGCACCCGTTGCTAGACATTCAACCATAAGAATGATCTTATCAAAAGCTGTACAAGATGATTTAAAGATTATTCAGCTAGACATTCCCACTGCATTCCTAAATGGAAAATTAAAGTCTGATGTATTTATCAAAATACCCCAAGGATTAGAAATAAAAAATAATAATACAGCATTAAAACTAAAGAGAGCATTGTATGGATTGGTTGAATCACCAAAATGTTGGAATGAAACAATTGACGAATTTGCGAAAGCTAAGGGATTCCAAAGATCAGAATATGATTACTGTTTGTATTACAAAGAAAATTGCTGGATGCTGATTTACGTGGACGATATCCTAATCATTGGAAACGGGGAAGATATAATTAAAGCACTTAAAGAGGAGTTTAACGCAAAATATATGGGCGAAATGAAGAACTTTCTCGGAATGGAAGTTAAAAGAAATAAAGATACATTAGAACTGAAACAAACTAAAATGATTGAAAAGATATTGACTAAATTCAAAATGGATGAATGTAACGGAATTAAAACACACATGGAAAGAAATTACAATATAGAAATAAAGGACAATACAGAAGTTTTGGACGTACCGTATCGAGAACTAATAGGAAGTCTAATTTATCTTACTGTAACAACAAGACCGGATTTAACATTTGCAGTGTCATATCTGAGCAGATTTTTAGATAAACCGAACGAAGAATTATGGAAAGCTGGAAAAAGAGAGGAGTGTTGA
Protein
MTAEMLHTDIAGPIRHQGPNGEKYFMTIIDDFSHFCVVYPLKSKSEATERLMNYVLRIENETGNKVKRIRCDNGGEYTANSLKLFCINRGISLEYTLPYTPQLNGISERMNRTLCDKTRTLLAETNLPKHLWCEAIQCAVYQLNRSPSWAINFKTPSGIFFGKNDLSRLRVFGSKAWATIVPKPDKLAKRAKETRMVGYGTVGYRLWDPEEDKIVISRDVRFDESDIKFKNNEEVKQVRYVQDKEEKINKEKEDIVEQEDEFKDAEMNTDQIEEAIIRPKRTINPPAYLKDYETNIENINETNIVICLSANAPETFEEAIKEPEWKNAINKELDALQRLQTWEEQEITDGRKLIDTKWIFKQKEDGTKKARLVARGFQQKKEDIFDSVYAPVARHSTIRMILSKAVQDDLKIIQLDIPTAFLNGKLKSDVFIKIPQGLEIKNNNTALKLKRALYGLVESPKCWNETIDEFAKAKGFQRSEYDYCLYYKENCWMLIYVDDILIIGNGEDIIKALKEEFNAKYMGEMKNFLGMEVKRNKDTLELKQTKMIEKILTKFKMDECNGIKTHMERNYNIEIKDNTEVLDVPYRELIGSLIYLTVTTRPDLTFAVSYLSRFLDKPNEELWKAGKREEC
Summary
Uniprot
O96968
A0A0A9XG27
A0A0A9Z0I6
A0A146KTS1
A0A1B6DFK3
A0A0A9WLY1
+ More
A0A146LV28 A0A224XA26 A0A0T6AT79 A0A3S2LQS6 A0A1Y1L115 A0A1Y1MQF0 A0A2J7QL79 A0A1Y1N9F7 A0A1Y1NCE2 A0A1Y1MJE9 A0A2R6RYD4 A0A182G1G7 A0A1B6F1U1 A0A182GA29 A0A0J7K689 A0A1B6C2D3 A0A1W7R6A3 B8YLY4 J9KJ72 A0A151RYL1 B8YLY5 B8YLY3 A0A151RCT9 B8YLY6 A0A2R6QHR4 A0A151R6U8 A0A2N9FQJ1 A0A2N9JB15 B8YLY7 A0A2N9GRZ0 A0A2N9EM80 A0A2N9I6N4 A0A2N9I1L5 A0A2N9FNY4 A0A2N9GHK9 A0A2N9FMR2 A0A2N9GLP2 H6U1F0 A0A151R2G6 A0A2N9ELY1 B1N668 A0A151TKT1 A0A087SVE4 A0A2N9H0T6 A0A2N9FV67 A0A151SDY6 A0A2N9ETC8 A0A2N9H6B2 A0A1Y1JXU0 H6U1E9 A0A2N9GY85 A0A2N9EYI8 A0A2N9HLU0 A0A151RJN4 A0A2N9HZR4 A0A2N9H4V1 A0A438DJP4 A0A438JKL2 A0A146L7E6 A0A438CJT7 A0A438D407 A0A438GBI6 A0A182GGE6 A0A438JB67 A0A438F4Z1 Q53ND1 A0A2N9HKJ4 A0A445B365 A0A2N9I0Q5 A0A2N9FSJ8 A0A251VAZ6 A0A2N9IN51 A0A438JSJ2 A0A438H9V6 Q2QQ81 Q53QA4 A0A2N9GA80 A0A2N9IUS6 A0A438IVY1 A0A2N9H171 A0A2N9I9F7 A0A438FRF7 A0A2N9IGN3 Q6AUC7 A0A438GNT7 A0A2N9HZ75 X1WUQ9 A0A438H767 A0A438DYX7 A5CBM1 A0A2K3N1R3 A0A251V331 A0A438HG45
A0A146LV28 A0A224XA26 A0A0T6AT79 A0A3S2LQS6 A0A1Y1L115 A0A1Y1MQF0 A0A2J7QL79 A0A1Y1N9F7 A0A1Y1NCE2 A0A1Y1MJE9 A0A2R6RYD4 A0A182G1G7 A0A1B6F1U1 A0A182GA29 A0A0J7K689 A0A1B6C2D3 A0A1W7R6A3 B8YLY4 J9KJ72 A0A151RYL1 B8YLY5 B8YLY3 A0A151RCT9 B8YLY6 A0A2R6QHR4 A0A151R6U8 A0A2N9FQJ1 A0A2N9JB15 B8YLY7 A0A2N9GRZ0 A0A2N9EM80 A0A2N9I6N4 A0A2N9I1L5 A0A2N9FNY4 A0A2N9GHK9 A0A2N9FMR2 A0A2N9GLP2 H6U1F0 A0A151R2G6 A0A2N9ELY1 B1N668 A0A151TKT1 A0A087SVE4 A0A2N9H0T6 A0A2N9FV67 A0A151SDY6 A0A2N9ETC8 A0A2N9H6B2 A0A1Y1JXU0 H6U1E9 A0A2N9GY85 A0A2N9EYI8 A0A2N9HLU0 A0A151RJN4 A0A2N9HZR4 A0A2N9H4V1 A0A438DJP4 A0A438JKL2 A0A146L7E6 A0A438CJT7 A0A438D407 A0A438GBI6 A0A182GGE6 A0A438JB67 A0A438F4Z1 Q53ND1 A0A2N9HKJ4 A0A445B365 A0A2N9I0Q5 A0A2N9FSJ8 A0A251VAZ6 A0A2N9IN51 A0A438JSJ2 A0A438H9V6 Q2QQ81 Q53QA4 A0A2N9GA80 A0A2N9IUS6 A0A438IVY1 A0A2N9H171 A0A2N9I9F7 A0A438FRF7 A0A2N9IGN3 Q6AUC7 A0A438GNT7 A0A2N9HZ75 X1WUQ9 A0A438H767 A0A438DYX7 A5CBM1 A0A2K3N1R3 A0A251V331 A0A438HG45
Pubmed
EMBL
AB014676
BAA74713.1
GBHO01027564
JAG16040.1
GBHO01004832
JAG38772.1
+ More
GDHC01019310 JAP99318.1 GEDC01012841 GEDC01000503 JAS24457.1 JAS36795.1 GBHO01035173 JAG08431.1 GDHC01007058 JAQ11571.1 GFTR01008662 JAW07764.1 LJIG01022881 KRT78250.1 RSAL01000408 RVE41872.1 GEZM01072005 JAV65635.1 GEZM01028916 JAV86196.1 NEVH01013252 PNF29308.1 GEZM01009498 GEZM01009497 JAV94551.1 GEZM01009496 GEZM01009495 JAV94560.1 GEZM01032204 JAV84770.1 NKQK01000002 PSS35035.1 JXUM01136904 KQ568499 KXJ68982.1 GECZ01025604 JAS44165.1 JXUM01155348 KQ572178 KXJ68066.1 LBMM01012772 KMQ86003.1 GEDC01029908 JAS07390.1 GEHC01000959 JAV46686.1 FJ544853 ACL97384.1 ABLF02041793 KQ483522 KYP47640.1 FJ544854 ACL97385.1 FJ544851 ACL97383.1 KQ483843 KYP40337.1 FJ544856 ACL97386.1 NKQK01000016 PSS08126.1 KQ484038 KYP38095.1 OIVN01001052 SPC89229.1 OIVN01006466 SPD33659.1 FJ544857 ACL97387.1 OIVN01002256 SPD02034.1 OIVN01000189 SPC75922.1 OIVN01004879 SPD19723.1 OIVN01004651 SPD18552.1 OIVN01001308 SPC92446.1 OIVN01001931 SPC99063.1 OIVN01000991 OIVN01001835 SPC88370.1 SPC98114.1 OIVN01002055 SPD00174.1 JN402333 AFB73912.1 KQ484186 KYP36645.1 OIVN01000183 SPC75833.1 EF094939 EF094940 ABO36622.1 ABO36636.1 CM003607 KYP67664.1 KK112142 KFM56833.1 OIVN01002680 SPD05612.1 OIVN01001196 SPC91020.1 KQ483417 KYP53032.1 OIVN01000557 SPC82046.1 OIVN01002904 SPD07378.1 GEZM01098220 JAV53943.1 JN402332 AFB73911.1 OIVN01002516 SPD04274.1 OIVN01000404 SPC79669.1 OIVN01003683 SPD12935.1 KQ483702 KYP42748.1 OIVN01004420 SPD17230.1 OIVN01002854 SPD06945.1 QGNW01001602 RVW35576.1 QGNW01000038 RVX09487.1 GDHC01015473 JAQ03156.1 QGNW01002195 RVW23445.1 QGNW01001808 RVW30183.1 QGNW01000493 RVW69546.1 JXUM01011189 KQ560325 KXJ83029.1 QGNW01000052 RVX06204.1 QGNW01001119 RVW55023.1 AC134048 DP000010 AAX92941.1 ABA91548.1 OIVN01003569 SPD12194.1 SDMP01000010 RYR33127.1 OIVN01004867 SPD19656.1 OIVN01001130 SPC90232.1 CM007892 OTG31811.1 OIVN01006120 SPD25513.1 QGNW01000029 RVX11927.1 QGNW01000255 RVW81243.1 DP000011 ABA98656.1 AC116368 AAX92861.1 ABA92739.1 OIVN01001666 SPC96375.1 OIVN01006261 SPD29147.1 QGNW01000079 RVX00796.1 OIVN01002656 SPD05381.1 OIVN01005059 SPD20680.1 QGNW01000769 RVW62538.1 OIVN01005591 SPD23209.1 AC129717 AAT85194.1 QGNW01000381 RVW73873.1 OIVN01004379 SPD17029.1 ABLF02020302 QGNW01000267 RVW80348.1 QGNW01001457 RVW40577.1 AM489192 CAN71109.1 ASHM01015075 PNX96991.1 CM007893 OTG30017.1 QGNW01000228 RVW83440.1
GDHC01019310 JAP99318.1 GEDC01012841 GEDC01000503 JAS24457.1 JAS36795.1 GBHO01035173 JAG08431.1 GDHC01007058 JAQ11571.1 GFTR01008662 JAW07764.1 LJIG01022881 KRT78250.1 RSAL01000408 RVE41872.1 GEZM01072005 JAV65635.1 GEZM01028916 JAV86196.1 NEVH01013252 PNF29308.1 GEZM01009498 GEZM01009497 JAV94551.1 GEZM01009496 GEZM01009495 JAV94560.1 GEZM01032204 JAV84770.1 NKQK01000002 PSS35035.1 JXUM01136904 KQ568499 KXJ68982.1 GECZ01025604 JAS44165.1 JXUM01155348 KQ572178 KXJ68066.1 LBMM01012772 KMQ86003.1 GEDC01029908 JAS07390.1 GEHC01000959 JAV46686.1 FJ544853 ACL97384.1 ABLF02041793 KQ483522 KYP47640.1 FJ544854 ACL97385.1 FJ544851 ACL97383.1 KQ483843 KYP40337.1 FJ544856 ACL97386.1 NKQK01000016 PSS08126.1 KQ484038 KYP38095.1 OIVN01001052 SPC89229.1 OIVN01006466 SPD33659.1 FJ544857 ACL97387.1 OIVN01002256 SPD02034.1 OIVN01000189 SPC75922.1 OIVN01004879 SPD19723.1 OIVN01004651 SPD18552.1 OIVN01001308 SPC92446.1 OIVN01001931 SPC99063.1 OIVN01000991 OIVN01001835 SPC88370.1 SPC98114.1 OIVN01002055 SPD00174.1 JN402333 AFB73912.1 KQ484186 KYP36645.1 OIVN01000183 SPC75833.1 EF094939 EF094940 ABO36622.1 ABO36636.1 CM003607 KYP67664.1 KK112142 KFM56833.1 OIVN01002680 SPD05612.1 OIVN01001196 SPC91020.1 KQ483417 KYP53032.1 OIVN01000557 SPC82046.1 OIVN01002904 SPD07378.1 GEZM01098220 JAV53943.1 JN402332 AFB73911.1 OIVN01002516 SPD04274.1 OIVN01000404 SPC79669.1 OIVN01003683 SPD12935.1 KQ483702 KYP42748.1 OIVN01004420 SPD17230.1 OIVN01002854 SPD06945.1 QGNW01001602 RVW35576.1 QGNW01000038 RVX09487.1 GDHC01015473 JAQ03156.1 QGNW01002195 RVW23445.1 QGNW01001808 RVW30183.1 QGNW01000493 RVW69546.1 JXUM01011189 KQ560325 KXJ83029.1 QGNW01000052 RVX06204.1 QGNW01001119 RVW55023.1 AC134048 DP000010 AAX92941.1 ABA91548.1 OIVN01003569 SPD12194.1 SDMP01000010 RYR33127.1 OIVN01004867 SPD19656.1 OIVN01001130 SPC90232.1 CM007892 OTG31811.1 OIVN01006120 SPD25513.1 QGNW01000029 RVX11927.1 QGNW01000255 RVW81243.1 DP000011 ABA98656.1 AC116368 AAX92861.1 ABA92739.1 OIVN01001666 SPC96375.1 OIVN01006261 SPD29147.1 QGNW01000079 RVX00796.1 OIVN01002656 SPD05381.1 OIVN01005059 SPD20680.1 QGNW01000769 RVW62538.1 OIVN01005591 SPD23209.1 AC129717 AAT85194.1 QGNW01000381 RVW73873.1 OIVN01004379 SPD17029.1 ABLF02020302 QGNW01000267 RVW80348.1 QGNW01001457 RVW40577.1 AM489192 CAN71109.1 ASHM01015075 PNX96991.1 CM007893 OTG30017.1 QGNW01000228 RVW83440.1
Proteomes
PRIDE
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR039537 Retrotran_Ty1/copia-like
IPR001584 Integrase_cat-core
IPR013103 RVT_2
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR025724 GAG-pre-integrase_dom
IPR025314 DUF4219
IPR038717 Tc1-like_DDE_dom
IPR036291 NAD(P)-bd_dom_sf
IPR013740 Redoxin
IPR020904 Sc_DH/Rdtase_CS
IPR002347 SDR_fam
IPR036249 Thioredoxin-like_sf
IPR037944 PRX5-like
IPR004859 Put_53exo
IPR041412 Xrn1_helical
IPR027073 5_3_exoribonuclease
IPR007123 Gelsolin-like_dom
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR036180 Gelsolin-like_dom_sf
IPR012990 Sec23_24_beta_S
IPR032451 SMARCC_C
IPR039537 Retrotran_Ty1/copia-like
IPR001584 Integrase_cat-core
IPR013103 RVT_2
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR025724 GAG-pre-integrase_dom
IPR025314 DUF4219
IPR038717 Tc1-like_DDE_dom
IPR036291 NAD(P)-bd_dom_sf
IPR013740 Redoxin
IPR020904 Sc_DH/Rdtase_CS
IPR002347 SDR_fam
IPR036249 Thioredoxin-like_sf
IPR037944 PRX5-like
IPR004859 Put_53exo
IPR041412 Xrn1_helical
IPR027073 5_3_exoribonuclease
IPR007123 Gelsolin-like_dom
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR036180 Gelsolin-like_dom_sf
IPR012990 Sec23_24_beta_S
IPR032451 SMARCC_C
SUPFAM
Gene 3D
ProteinModelPortal
O96968
A0A0A9XG27
A0A0A9Z0I6
A0A146KTS1
A0A1B6DFK3
A0A0A9WLY1
+ More
A0A146LV28 A0A224XA26 A0A0T6AT79 A0A3S2LQS6 A0A1Y1L115 A0A1Y1MQF0 A0A2J7QL79 A0A1Y1N9F7 A0A1Y1NCE2 A0A1Y1MJE9 A0A2R6RYD4 A0A182G1G7 A0A1B6F1U1 A0A182GA29 A0A0J7K689 A0A1B6C2D3 A0A1W7R6A3 B8YLY4 J9KJ72 A0A151RYL1 B8YLY5 B8YLY3 A0A151RCT9 B8YLY6 A0A2R6QHR4 A0A151R6U8 A0A2N9FQJ1 A0A2N9JB15 B8YLY7 A0A2N9GRZ0 A0A2N9EM80 A0A2N9I6N4 A0A2N9I1L5 A0A2N9FNY4 A0A2N9GHK9 A0A2N9FMR2 A0A2N9GLP2 H6U1F0 A0A151R2G6 A0A2N9ELY1 B1N668 A0A151TKT1 A0A087SVE4 A0A2N9H0T6 A0A2N9FV67 A0A151SDY6 A0A2N9ETC8 A0A2N9H6B2 A0A1Y1JXU0 H6U1E9 A0A2N9GY85 A0A2N9EYI8 A0A2N9HLU0 A0A151RJN4 A0A2N9HZR4 A0A2N9H4V1 A0A438DJP4 A0A438JKL2 A0A146L7E6 A0A438CJT7 A0A438D407 A0A438GBI6 A0A182GGE6 A0A438JB67 A0A438F4Z1 Q53ND1 A0A2N9HKJ4 A0A445B365 A0A2N9I0Q5 A0A2N9FSJ8 A0A251VAZ6 A0A2N9IN51 A0A438JSJ2 A0A438H9V6 Q2QQ81 Q53QA4 A0A2N9GA80 A0A2N9IUS6 A0A438IVY1 A0A2N9H171 A0A2N9I9F7 A0A438FRF7 A0A2N9IGN3 Q6AUC7 A0A438GNT7 A0A2N9HZ75 X1WUQ9 A0A438H767 A0A438DYX7 A5CBM1 A0A2K3N1R3 A0A251V331 A0A438HG45
A0A146LV28 A0A224XA26 A0A0T6AT79 A0A3S2LQS6 A0A1Y1L115 A0A1Y1MQF0 A0A2J7QL79 A0A1Y1N9F7 A0A1Y1NCE2 A0A1Y1MJE9 A0A2R6RYD4 A0A182G1G7 A0A1B6F1U1 A0A182GA29 A0A0J7K689 A0A1B6C2D3 A0A1W7R6A3 B8YLY4 J9KJ72 A0A151RYL1 B8YLY5 B8YLY3 A0A151RCT9 B8YLY6 A0A2R6QHR4 A0A151R6U8 A0A2N9FQJ1 A0A2N9JB15 B8YLY7 A0A2N9GRZ0 A0A2N9EM80 A0A2N9I6N4 A0A2N9I1L5 A0A2N9FNY4 A0A2N9GHK9 A0A2N9FMR2 A0A2N9GLP2 H6U1F0 A0A151R2G6 A0A2N9ELY1 B1N668 A0A151TKT1 A0A087SVE4 A0A2N9H0T6 A0A2N9FV67 A0A151SDY6 A0A2N9ETC8 A0A2N9H6B2 A0A1Y1JXU0 H6U1E9 A0A2N9GY85 A0A2N9EYI8 A0A2N9HLU0 A0A151RJN4 A0A2N9HZR4 A0A2N9H4V1 A0A438DJP4 A0A438JKL2 A0A146L7E6 A0A438CJT7 A0A438D407 A0A438GBI6 A0A182GGE6 A0A438JB67 A0A438F4Z1 Q53ND1 A0A2N9HKJ4 A0A445B365 A0A2N9I0Q5 A0A2N9FSJ8 A0A251VAZ6 A0A2N9IN51 A0A438JSJ2 A0A438H9V6 Q2QQ81 Q53QA4 A0A2N9GA80 A0A2N9IUS6 A0A438IVY1 A0A2N9H171 A0A2N9I9F7 A0A438FRF7 A0A2N9IGN3 Q6AUC7 A0A438GNT7 A0A2N9HZ75 X1WUQ9 A0A438H767 A0A438DYX7 A5CBM1 A0A2K3N1R3 A0A251V331 A0A438HG45
PDB
3DLR
E-value=1.31631e-06,
Score=127
Ontologies
GO
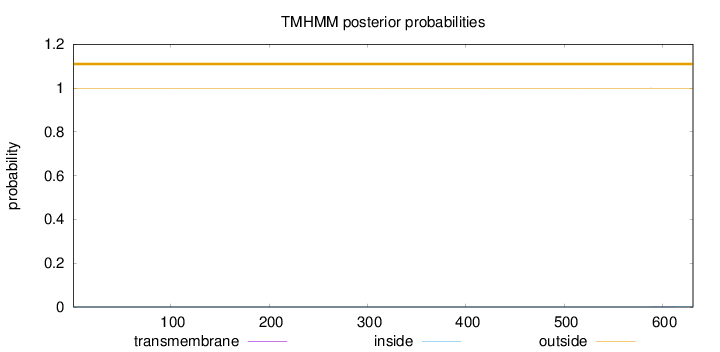
Topology
Length:
631
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01454
Exp number, first 60 AAs:
0.00077
Total prob of N-in:
0.00057
outside
1 - 631
Population Genetic Test Statistics
Pi
197.94814
Theta
192.785606
Tajima's D
-0.683659
CLR
206.615273
CSRT
0.196390180490975
Interpretation
Uncertain
