Gene
KWMTBOMO08248
Pre Gene Modal
BGIBMGA009518
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_alpha-N-acetylgalactosaminidase-like_[Bombyx_mori]
Full name
Alpha-galactosidase
Location in the cell
Extracellular Reliability : 1.062 Lysosomal Reliability : 1.663 PlasmaMembrane Reliability : 1.015
Sequence
CDS
ATGGCTTCGTGGAAGTTTATCGCTGTGTCGACGGTTTGTTTGTTGGTGTTTCTGTCGGGTAACATCTCGGGACTGGACAATGGCTTGGTGCTGACGCCGCCCATGGGCTGGCTGGCCTGGGAACGGTTTAGGTGTAACACCGATTGCAAGAATGACCCTGAAAATTGTATCAGCGATCGTCTATTCCGTACCATGACCGATATCTTAGTGACCGAAGGGTATACCGCGGCTGGATATGAGTACATCAACGTGGACGACTGCTGGCTGGAGCGGGAGAGGGACGAGAGAGGTAGACTAGTGCCGGACAGAGAACGGTTCCCTTACGGAATGAAGAGCTTAGCCGATTATGTACACAGCAAGGGTCTGAAATTCGGTATTTACGAGGACTACGGTAATTTCACTTGCGCTGGCTACCCCGGAGTCGTCGGCCATCTTTCCGGAGATGCGACAACCTTCGCCTCATGGGGCGTCGACTACGTGAAGCTGGACGGGTGCTACGCCCTGCCAACGGACATGGATTACGGCTACCCTGAGTTCGGGAGACAGCTCAATCTTACTGGTCGTCAGATGGTGTACTCCTGTAGTTGGCCCGTCTATCAGATCTACGCTGGTCTACAGCCAAATTTTTCGTCAATAATCGAGCACTGCAATCTGTGGCGTAACTTCGACGACATCCAGGACTCGTGGGCCTCCGTCGAGTCTATCATAGACTACTATGGAAACCACCAAGACGTCATCGTGCCCAACGCTGGGCCGGGACACTGGAACGACCCCGACATGTTGATCGTCGGCAACTTTGGACTATCGTATGAGCAGAGCAAGACGCAGTTCGCACTATGGGCGATCCTGGCCGCGCCGCTGCTGATGAGCGTCGACCTGCGCACCATCAGGCCGGAGTACAAGGCCATTCTGCAGAACAGGAAGATCATCGAGGTCGACCAGGACCCCCTCGGCATACAAGGCAGGAGGATCTACAAGCACCGCGGCATCGAAATCTGGTCCCGCCCCATCCTGCCCATCCAGCAGGGACAGTTCTACTCGTACGCCGTGGCCTTCGTCAACAGGAGGACAGACGGGACTCCTTCCGACGTCGCCGTCACGCTGAGGGAGCTGGGGCTCAGCAACCCCACCGGCTACAGGGTCGAGGACCTGTACGAGGACGTGGATTACGGCGTGCTGTCGCCGCAGACGAAGATCAAGGTCAAGGTCAACCCCTCAGGTGTGGTGATTTTGAGGGCAGACGCGCAACCGGGCTACGCTTTCAACGCGATAACGACGCGGACTCCGTACACGCCACTCAACGACGTGTTCCGACTGACCAAGAAATAG
Protein
MASWKFIAVSTVCLLVFLSGNISGLDNGLVLTPPMGWLAWERFRCNTDCKNDPENCISDRLFRTMTDILVTEGYTAAGYEYINVDDCWLERERDERGRLVPDRERFPYGMKSLADYVHSKGLKFGIYEDYGNFTCAGYPGVVGHLSGDATTFASWGVDYVKLDGCYALPTDMDYGYPEFGRQLNLTGRQMVYSCSWPVYQIYAGLQPNFSSIIEHCNLWRNFDDIQDSWASVESIIDYYGNHQDVIVPNAGPGHWNDPDMLIVGNFGLSYEQSKTQFALWAILAAPLLMSVDLRTIRPEYKAILQNRKIIEVDQDPLGIQGRRIYKHRGIEIWSRPILPIQQGQFYSYAVAFVNRRTDGTPSDVAVTLRELGLSNPTGYRVEDLYEDVDYGVLSPQTKIKVKVNPSGVVILRADAQPGYAFNAITTRTPYTPLNDVFRLTKK
Summary
Subunit
Homodimer.
Similarity
Belongs to the glycosyl hydrolase 27 family.
Feature
chain Alpha-galactosidase
Uniprot
A0A437BPU5
A0A2W1BQY8
A0A2A4JJP4
S4NSM8
I4DIL5
A0A0N1PES9
+ More
E2B637 F4WJD6 A0A026WYH3 A0A151J0R5 D1ZZP5 J3JUA1 K7J2S2 A0A1Y1M843 A0A232EJJ6 A0A0J7KXZ7 U4UPP5 E2A7Y1 A0A1J1IRP1 Q17A03 A0A1B6CLQ8 A0A182Q255 A0A2A3EAC4 A0A1Q3G433 A0A195CCG6 A0A1Q3G451 A0A1S4H2E7 A0A182ICP7 A0A151WST7 A0A084VNI9 A0A1L8DR08 A0A182MV14 A0A195BWB5 A0A195FQA7 A0A1L8DQP1 A0A182VNL1 A0A182IPD0 A0A182Y8D8 A0A1L8DQU5 Q7Q1V0 A0A182L619 A0A1L8DQP5 A0A0L7QQI1 A0A2M4DMN4 A0A2M3Z177 A0A2M4AC36 A0A2M4ACK0 A0A2M4BML2 A0A2M4DMS1 A0A2M4DMQ3 A0A158NE94 E0VBI5 A0A0K8SY61 A0A336M9C7 A0A0A9VX47 A0A2J7QEQ5 D2T1A8 A0A3L8DBX5 E9IEL9 A0A0M8ZS78 A0A411G871 T1HJA0 A0A154PHX5 A0A023F4E1 B3MPM5 A0A2S2ND41 J9JRF8 A0A310SAX6 B4HWH1 B4Q8Q4 B3N935 Q9VL27 Q8MYY3 A0A1W4UUS4 B4NZ59 B4JP37 B4KHN3 B4G7B6 A0A2S2PVG7 A0A067RV68 A0A0C9R4U7 A0A1A9ZVN6 A0A1A9UT81 A0A3B0K177 A0A023ETE0 A0A3B0K9B0 A0A1A9WLA3 B4MUD8 A0A1A9Y6Y0 B4M8H2 B5DHX9 A0A088A332 A0A0K2T4T3 A0A182F2Q2 A0A182K027 A0A1S3DTP6 A0A182NB11 B0WEB8 A0A182RCD2 A0A1B0CBE8
E2B637 F4WJD6 A0A026WYH3 A0A151J0R5 D1ZZP5 J3JUA1 K7J2S2 A0A1Y1M843 A0A232EJJ6 A0A0J7KXZ7 U4UPP5 E2A7Y1 A0A1J1IRP1 Q17A03 A0A1B6CLQ8 A0A182Q255 A0A2A3EAC4 A0A1Q3G433 A0A195CCG6 A0A1Q3G451 A0A1S4H2E7 A0A182ICP7 A0A151WST7 A0A084VNI9 A0A1L8DR08 A0A182MV14 A0A195BWB5 A0A195FQA7 A0A1L8DQP1 A0A182VNL1 A0A182IPD0 A0A182Y8D8 A0A1L8DQU5 Q7Q1V0 A0A182L619 A0A1L8DQP5 A0A0L7QQI1 A0A2M4DMN4 A0A2M3Z177 A0A2M4AC36 A0A2M4ACK0 A0A2M4BML2 A0A2M4DMS1 A0A2M4DMQ3 A0A158NE94 E0VBI5 A0A0K8SY61 A0A336M9C7 A0A0A9VX47 A0A2J7QEQ5 D2T1A8 A0A3L8DBX5 E9IEL9 A0A0M8ZS78 A0A411G871 T1HJA0 A0A154PHX5 A0A023F4E1 B3MPM5 A0A2S2ND41 J9JRF8 A0A310SAX6 B4HWH1 B4Q8Q4 B3N935 Q9VL27 Q8MYY3 A0A1W4UUS4 B4NZ59 B4JP37 B4KHN3 B4G7B6 A0A2S2PVG7 A0A067RV68 A0A0C9R4U7 A0A1A9ZVN6 A0A1A9UT81 A0A3B0K177 A0A023ETE0 A0A3B0K9B0 A0A1A9WLA3 B4MUD8 A0A1A9Y6Y0 B4M8H2 B5DHX9 A0A088A332 A0A0K2T4T3 A0A182F2Q2 A0A182K027 A0A1S3DTP6 A0A182NB11 B0WEB8 A0A182RCD2 A0A1B0CBE8
EC Number
3.2.1.-
Pubmed
28756777
23622113
22651552
26354079
20798317
21719571
+ More
24508170 18362917 19820115 22516182 20075255 28004739 28648823 23537049 17510324 12364791 24438588 25244985 20966253 21347285 20566863 25401762 30249741 21282665 25474469 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24845553 24945155 15632085 23185243
24508170 18362917 19820115 22516182 20075255 28004739 28648823 23537049 17510324 12364791 24438588 25244985 20966253 21347285 20566863 25401762 30249741 21282665 25474469 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24845553 24945155 15632085 23185243
EMBL
RSAL01000021
RVE52412.1
KZ149927
PZC77532.1
NWSH01001168
PCG72287.1
+ More
GAIX01014012 JAA78548.1 AK401133 BAM17755.1 KQ458751 KPJ05313.1 GL445914 EFN88826.1 GL888182 EGI65714.1 KK107079 EZA60164.1 KQ980636 KYN14937.1 KQ971338 EFA01818.1 BT126814 AEE61776.1 GEZM01037887 JAV81864.1 NNAY01003983 OXU18533.1 LBMM01002203 KMQ95149.1 KB632308 ERL91995.1 GL437442 EFN70460.1 CVRI01000058 CRL02887.1 CH477342 EAT43068.1 GEDC01029663 GEDC01022894 JAS07635.1 JAS14404.1 AXCN02000647 KZ288322 PBC28159.1 GFDL01000485 JAV34560.1 KQ978023 KYM97921.1 GFDL01000468 JAV34577.1 AAAB01008980 APCN01005736 KQ982769 KYQ50857.1 ATLV01014759 KE524984 KFB39533.1 GFDF01005314 JAV08770.1 AXCM01001912 KQ976401 KYM92575.1 KQ981335 KYN42658.1 GFDF01005311 JAV08773.1 GFDF01005313 JAV08771.1 EAA14548.3 GFDF01005312 JAV08772.1 KQ414786 KOC60877.1 GGFL01014652 MBW78830.1 GGFM01001531 MBW22282.1 GGFK01004999 MBW38320.1 GGFK01005007 MBW38328.1 GGFJ01005176 MBW54317.1 GGFL01014653 MBW78831.1 GGFL01014654 MBW78832.1 ADTU01013175 DS235030 EEB10741.1 GBRD01007725 JAG58096.1 UFQS01000272 UFQT01000272 SSX02272.1 SSX22648.1 GBHO01043838 JAF99765.1 NEVH01015307 PNF27047.1 FM986353 CAX20733.1 QOIP01000010 RLU17408.1 GL762647 EFZ20978.1 KQ435878 KOX70065.1 MH366297 QBB02106.1 ACPB03006489 KQ434896 KZC10800.1 GBBI01002331 JAC16381.1 CH902620 EDV32273.1 GGMR01002419 MBY15038.1 ABLF02027051 ABLF02027053 KQ765605 OAD53851.1 CH480818 EDW52366.1 CM000361 CM002910 EDX04469.1 KMY89418.1 CH954177 EDV58470.2 AE014134 BT044084 AAF52871.2 ACH92149.1 AY113489 AAM29494.1 CM000157 EDW88754.1 CH916372 EDV99462.1 CH933807 EDW12312.1 CH479180 EDW28370.1 GGMS01000258 MBY69461.1 KK852444 KDR23774.1 GBYB01007777 JAG77544.1 OUUW01000006 SPP81610.1 GAPW01001944 JAC11654.1 SPP81611.1 CH963857 EDW76064.1 CH940654 EDW57498.2 CH379059 EDY69906.1 KRT03770.1 HACA01003085 CDW20446.1 DS231907 EDS45478.1 AJWK01005088
GAIX01014012 JAA78548.1 AK401133 BAM17755.1 KQ458751 KPJ05313.1 GL445914 EFN88826.1 GL888182 EGI65714.1 KK107079 EZA60164.1 KQ980636 KYN14937.1 KQ971338 EFA01818.1 BT126814 AEE61776.1 GEZM01037887 JAV81864.1 NNAY01003983 OXU18533.1 LBMM01002203 KMQ95149.1 KB632308 ERL91995.1 GL437442 EFN70460.1 CVRI01000058 CRL02887.1 CH477342 EAT43068.1 GEDC01029663 GEDC01022894 JAS07635.1 JAS14404.1 AXCN02000647 KZ288322 PBC28159.1 GFDL01000485 JAV34560.1 KQ978023 KYM97921.1 GFDL01000468 JAV34577.1 AAAB01008980 APCN01005736 KQ982769 KYQ50857.1 ATLV01014759 KE524984 KFB39533.1 GFDF01005314 JAV08770.1 AXCM01001912 KQ976401 KYM92575.1 KQ981335 KYN42658.1 GFDF01005311 JAV08773.1 GFDF01005313 JAV08771.1 EAA14548.3 GFDF01005312 JAV08772.1 KQ414786 KOC60877.1 GGFL01014652 MBW78830.1 GGFM01001531 MBW22282.1 GGFK01004999 MBW38320.1 GGFK01005007 MBW38328.1 GGFJ01005176 MBW54317.1 GGFL01014653 MBW78831.1 GGFL01014654 MBW78832.1 ADTU01013175 DS235030 EEB10741.1 GBRD01007725 JAG58096.1 UFQS01000272 UFQT01000272 SSX02272.1 SSX22648.1 GBHO01043838 JAF99765.1 NEVH01015307 PNF27047.1 FM986353 CAX20733.1 QOIP01000010 RLU17408.1 GL762647 EFZ20978.1 KQ435878 KOX70065.1 MH366297 QBB02106.1 ACPB03006489 KQ434896 KZC10800.1 GBBI01002331 JAC16381.1 CH902620 EDV32273.1 GGMR01002419 MBY15038.1 ABLF02027051 ABLF02027053 KQ765605 OAD53851.1 CH480818 EDW52366.1 CM000361 CM002910 EDX04469.1 KMY89418.1 CH954177 EDV58470.2 AE014134 BT044084 AAF52871.2 ACH92149.1 AY113489 AAM29494.1 CM000157 EDW88754.1 CH916372 EDV99462.1 CH933807 EDW12312.1 CH479180 EDW28370.1 GGMS01000258 MBY69461.1 KK852444 KDR23774.1 GBYB01007777 JAG77544.1 OUUW01000006 SPP81610.1 GAPW01001944 JAC11654.1 SPP81611.1 CH963857 EDW76064.1 CH940654 EDW57498.2 CH379059 EDY69906.1 KRT03770.1 HACA01003085 CDW20446.1 DS231907 EDS45478.1 AJWK01005088
Proteomes
UP000283053
UP000218220
UP000053268
UP000008237
UP000007755
UP000053097
+ More
UP000078492 UP000007266 UP000002358 UP000215335 UP000036403 UP000030742 UP000000311 UP000183832 UP000008820 UP000075886 UP000242457 UP000078542 UP000075840 UP000075809 UP000030765 UP000075883 UP000078540 UP000078541 UP000075903 UP000075880 UP000076408 UP000007062 UP000075882 UP000053825 UP000005205 UP000009046 UP000235965 UP000279307 UP000053105 UP000015103 UP000076502 UP000007801 UP000007819 UP000001292 UP000000304 UP000008711 UP000000803 UP000192221 UP000002282 UP000001070 UP000009192 UP000008744 UP000027135 UP000092445 UP000078200 UP000268350 UP000091820 UP000007798 UP000092443 UP000008792 UP000001819 UP000005203 UP000069272 UP000075881 UP000079169 UP000075884 UP000002320 UP000075900 UP000092461
UP000078492 UP000007266 UP000002358 UP000215335 UP000036403 UP000030742 UP000000311 UP000183832 UP000008820 UP000075886 UP000242457 UP000078542 UP000075840 UP000075809 UP000030765 UP000075883 UP000078540 UP000078541 UP000075903 UP000075880 UP000076408 UP000007062 UP000075882 UP000053825 UP000005205 UP000009046 UP000235965 UP000279307 UP000053105 UP000015103 UP000076502 UP000007801 UP000007819 UP000001292 UP000000304 UP000008711 UP000000803 UP000192221 UP000002282 UP000001070 UP000009192 UP000008744 UP000027135 UP000092445 UP000078200 UP000268350 UP000091820 UP000007798 UP000092443 UP000008792 UP000001819 UP000005203 UP000069272 UP000075881 UP000079169 UP000075884 UP000002320 UP000075900 UP000092461
PRIDE
Interpro
ProteinModelPortal
A0A437BPU5
A0A2W1BQY8
A0A2A4JJP4
S4NSM8
I4DIL5
A0A0N1PES9
+ More
E2B637 F4WJD6 A0A026WYH3 A0A151J0R5 D1ZZP5 J3JUA1 K7J2S2 A0A1Y1M843 A0A232EJJ6 A0A0J7KXZ7 U4UPP5 E2A7Y1 A0A1J1IRP1 Q17A03 A0A1B6CLQ8 A0A182Q255 A0A2A3EAC4 A0A1Q3G433 A0A195CCG6 A0A1Q3G451 A0A1S4H2E7 A0A182ICP7 A0A151WST7 A0A084VNI9 A0A1L8DR08 A0A182MV14 A0A195BWB5 A0A195FQA7 A0A1L8DQP1 A0A182VNL1 A0A182IPD0 A0A182Y8D8 A0A1L8DQU5 Q7Q1V0 A0A182L619 A0A1L8DQP5 A0A0L7QQI1 A0A2M4DMN4 A0A2M3Z177 A0A2M4AC36 A0A2M4ACK0 A0A2M4BML2 A0A2M4DMS1 A0A2M4DMQ3 A0A158NE94 E0VBI5 A0A0K8SY61 A0A336M9C7 A0A0A9VX47 A0A2J7QEQ5 D2T1A8 A0A3L8DBX5 E9IEL9 A0A0M8ZS78 A0A411G871 T1HJA0 A0A154PHX5 A0A023F4E1 B3MPM5 A0A2S2ND41 J9JRF8 A0A310SAX6 B4HWH1 B4Q8Q4 B3N935 Q9VL27 Q8MYY3 A0A1W4UUS4 B4NZ59 B4JP37 B4KHN3 B4G7B6 A0A2S2PVG7 A0A067RV68 A0A0C9R4U7 A0A1A9ZVN6 A0A1A9UT81 A0A3B0K177 A0A023ETE0 A0A3B0K9B0 A0A1A9WLA3 B4MUD8 A0A1A9Y6Y0 B4M8H2 B5DHX9 A0A088A332 A0A0K2T4T3 A0A182F2Q2 A0A182K027 A0A1S3DTP6 A0A182NB11 B0WEB8 A0A182RCD2 A0A1B0CBE8
E2B637 F4WJD6 A0A026WYH3 A0A151J0R5 D1ZZP5 J3JUA1 K7J2S2 A0A1Y1M843 A0A232EJJ6 A0A0J7KXZ7 U4UPP5 E2A7Y1 A0A1J1IRP1 Q17A03 A0A1B6CLQ8 A0A182Q255 A0A2A3EAC4 A0A1Q3G433 A0A195CCG6 A0A1Q3G451 A0A1S4H2E7 A0A182ICP7 A0A151WST7 A0A084VNI9 A0A1L8DR08 A0A182MV14 A0A195BWB5 A0A195FQA7 A0A1L8DQP1 A0A182VNL1 A0A182IPD0 A0A182Y8D8 A0A1L8DQU5 Q7Q1V0 A0A182L619 A0A1L8DQP5 A0A0L7QQI1 A0A2M4DMN4 A0A2M3Z177 A0A2M4AC36 A0A2M4ACK0 A0A2M4BML2 A0A2M4DMS1 A0A2M4DMQ3 A0A158NE94 E0VBI5 A0A0K8SY61 A0A336M9C7 A0A0A9VX47 A0A2J7QEQ5 D2T1A8 A0A3L8DBX5 E9IEL9 A0A0M8ZS78 A0A411G871 T1HJA0 A0A154PHX5 A0A023F4E1 B3MPM5 A0A2S2ND41 J9JRF8 A0A310SAX6 B4HWH1 B4Q8Q4 B3N935 Q9VL27 Q8MYY3 A0A1W4UUS4 B4NZ59 B4JP37 B4KHN3 B4G7B6 A0A2S2PVG7 A0A067RV68 A0A0C9R4U7 A0A1A9ZVN6 A0A1A9UT81 A0A3B0K177 A0A023ETE0 A0A3B0K9B0 A0A1A9WLA3 B4MUD8 A0A1A9Y6Y0 B4M8H2 B5DHX9 A0A088A332 A0A0K2T4T3 A0A182F2Q2 A0A182K027 A0A1S3DTP6 A0A182NB11 B0WEB8 A0A182RCD2 A0A1B0CBE8
PDB
1KTC
E-value=8.81748e-127,
Score=1162
Ontologies
PATHWAY
GO
PANTHER
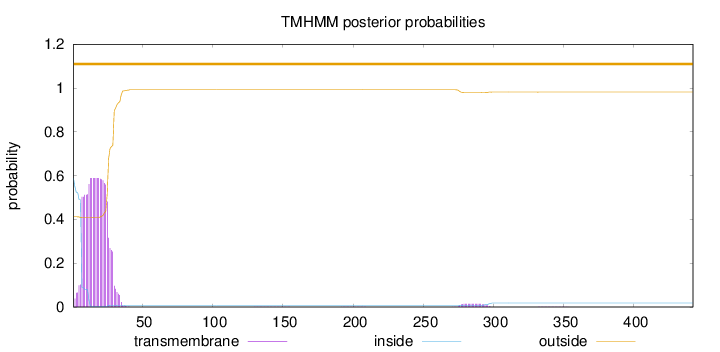
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.980913

Length:
442
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.7427
Exp number, first 60 AAs:
12.45315
Total prob of N-in:
0.58749
POSSIBLE N-term signal
sequence
outside
1 - 442
Population Genetic Test Statistics
Pi
267.802343
Theta
158.840899
Tajima's D
2.160061
CLR
0.238835
CSRT
0.903054847257637
Interpretation
Uncertain
