Gene
KWMTBOMO08240
Pre Gene Modal
BGIBMGA009453
Annotation
PREDICTED:_AN1-type_zinc_finger_protein_2A-like_[Papilio_polytes]
Location in the cell
Extracellular Reliability : 2.348 Nuclear Reliability : 2.178
Sequence
CDS
ATGGAATTTCCTCATCTTGGAAAAAATTGTTCTTATGAATCTTGCAATAAACTGGATTTCTTGCCGATGAAATGTGGTGCTTGTCGTGAAGTGTTCTGCGCCGATCATTTTGCTTATGCGAAACACGAGTGCCCCGAGCCGAACGTACGTGACGTGCAAGTGCCAGTGTGTCCGCTGTGCGGGGCTCCAGTGCCGGGTCGGAGGGGTCAACCACCAGACCTGGCAGTTGGGGCTCACATAGATAACCAGTGTACATCTGATCCTGCAGTAGAGAGACGCAAGAAGGTGTTCACGAACCGTTGTTCATACAAGAGCTGCAAAAGGAAGGAGATGGTGCCGCTGGTATGTTCTGAATGCGCACTCAACTTCTGTCTAAGGCACAGACACACGGCCGACCACTTGTGCGAAGGGAAACTTGCCGCCAAGAGGAGACAAGCCGCTAAGGCTGCCATGGCCCGGATGAGACAAAACGAAAGTCTATGGATCCGGAACGAGGCCCCGCTGCCCTCCGCTGTCAGCAGCTTCTCTGAGGTCCAGGGGAACATGACAGAAGACGAGGCGTTGGCTCACGCGCTGGCGTTGTCGATGCAGGAGAACGTGAACATAGAGTCCGGCGACACCAGCCTCGCGCGCGCCCTCACCCGCCAGCGCGTCGGCGGCGACCAGCCCGCCCGCTGCACCGTGTCCTAA
Protein
MEFPHLGKNCSYESCNKLDFLPMKCGACREVFCADHFAYAKHECPEPNVRDVQVPVCPLCGAPVPGRRGQPPDLAVGAHIDNQCTSDPAVERRKKVFTNRCSYKSCKRKEMVPLVCSECALNFCLRHRHTADHLCEGKLAAKRRQAAKAAMARMRQNESLWIRNEAPLPSAVSSFSEVQGNMTEDEALAHALALSMQENVNIESGDTSLARALTRQRVGGDQPARCTVS
Summary
Uniprot
H9JIV2
A0A0N0PA85
A0A437BPS7
A0A2W1BWE1
A0A2A4JR49
A0A212ET10
+ More
A0A1E1W7M5 S4PTE2 A0A1E1W7I5 A0A1Y1KFR5 A0A1B6C7F1 A0A1S3D3G5 D2A2F5 A0A1B6HEE5 A0A084VE19 N6U5N9 A0A0K8W3Q9 A0A034WHM7 A0A182QTW0 B4KJL5 A0A0A1WKC5 A0A182RNL7 A0A182INA2 W5JGC7 A0A2M4BY54 A0A182LYQ0 W8CAZ0 A0A1Q3F2X5 A0A182VQ82 E5L9P2 A0A1L8EFL7 A0A182HTC8 A0A0P5TN19 E2BQ21 A0A0P5E104 A0A0P6CQU6 A0A0A9WM26 A0A2M4AUT6 B4N105 A0A182LFH9 A0A182XEI2 A0A182N4L7 A0A2M4AVA9 A0A182VN11 Q7QBW5 A0A1L8EG21 A0A0L0BVM4 A0A182TI46 A0A3B0JBU9 A0A1Q3F3S8 A0A182PTH4 A0A182Y7B0 A0A182GJX2 Q9VQQ7 A0A182K5M3 A0A1I8MPU0 A0A023EKP1 B4LSG3 B7QEH7 B4INK1 B4NXW6 B4Q9K3 A0A2C9LW43 A0A0P5VKA4 A0A069DXJ0 A0A0P4VIZ5 A0A2C9LW62 A0A0C9R2Z5 A0A131XXQ4 B4JBU5 A0A0J7KJ86 A0A0K8R7Z4 A0A0L7R0F8 A0A1A9WXK8 A0A1I8PVE6 Q16UF2 A0A1S4G260 A0A195CG29 U5ES23 A0A1B0CW47 A0A087UJK0 A0A0V0GD02 A0A1A9Z7W1 B3N321 A0A195BWK5 F6R192 A0A0K8TNQ6 A0A232ER66 A0A026W763 A0A1W4UUH4 A0A1S3IZ84 A0A1S3IZT3 A0A1S3J062 K7J1D4 Q0P3R6 A0A1S3IZL5 Q16G40
A0A1E1W7M5 S4PTE2 A0A1E1W7I5 A0A1Y1KFR5 A0A1B6C7F1 A0A1S3D3G5 D2A2F5 A0A1B6HEE5 A0A084VE19 N6U5N9 A0A0K8W3Q9 A0A034WHM7 A0A182QTW0 B4KJL5 A0A0A1WKC5 A0A182RNL7 A0A182INA2 W5JGC7 A0A2M4BY54 A0A182LYQ0 W8CAZ0 A0A1Q3F2X5 A0A182VQ82 E5L9P2 A0A1L8EFL7 A0A182HTC8 A0A0P5TN19 E2BQ21 A0A0P5E104 A0A0P6CQU6 A0A0A9WM26 A0A2M4AUT6 B4N105 A0A182LFH9 A0A182XEI2 A0A182N4L7 A0A2M4AVA9 A0A182VN11 Q7QBW5 A0A1L8EG21 A0A0L0BVM4 A0A182TI46 A0A3B0JBU9 A0A1Q3F3S8 A0A182PTH4 A0A182Y7B0 A0A182GJX2 Q9VQQ7 A0A182K5M3 A0A1I8MPU0 A0A023EKP1 B4LSG3 B7QEH7 B4INK1 B4NXW6 B4Q9K3 A0A2C9LW43 A0A0P5VKA4 A0A069DXJ0 A0A0P4VIZ5 A0A2C9LW62 A0A0C9R2Z5 A0A131XXQ4 B4JBU5 A0A0J7KJ86 A0A0K8R7Z4 A0A0L7R0F8 A0A1A9WXK8 A0A1I8PVE6 Q16UF2 A0A1S4G260 A0A195CG29 U5ES23 A0A1B0CW47 A0A087UJK0 A0A0V0GD02 A0A1A9Z7W1 B3N321 A0A195BWK5 F6R192 A0A0K8TNQ6 A0A232ER66 A0A026W763 A0A1W4UUH4 A0A1S3IZ84 A0A1S3IZT3 A0A1S3J062 K7J1D4 Q0P3R6 A0A1S3IZL5 Q16G40
Pubmed
19121390
26354079
28756777
22118469
23622113
28004739
+ More
18362917 19820115 24438588 23537049 25348373 17994087 25830018 20920257 23761445 24495485 20978910 20798317 25401762 26823975 20966253 12364791 14747013 17210077 26108605 25244985 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24945155 17550304 22936249 15562597 26334808 27129103 17510324 12481130 15114417 26369729 28648823 24508170 30249741 20075255 27762356
18362917 19820115 24438588 23537049 25348373 17994087 25830018 20920257 23761445 24495485 20978910 20798317 25401762 26823975 20966253 12364791 14747013 17210077 26108605 25244985 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24945155 17550304 22936249 15562597 26334808 27129103 17510324 12481130 15114417 26369729 28648823 24508170 30249741 20075255 27762356
EMBL
BABH01023139
KQ458751
KPJ05320.1
RSAL01000021
RVE52403.1
KZ149927
+ More
PZC77527.1 NWSH01000813 PCG74084.1 AGBW02012702 OWR44584.1 GDQN01008790 GDQN01008107 JAT82264.1 JAT82947.1 GAIX01012193 JAA80367.1 GDQN01008121 JAT82933.1 GEZM01084997 GEZM01084996 JAV60329.1 GEDC01027896 JAS09402.1 KQ971338 EFA02191.1 GECU01034735 JAS72971.1 ATLV01012143 KE524764 KFB36213.1 APGK01039000 KB740966 KB632186 ENN76930.1 ERL89700.1 GDHF01006501 JAI45813.1 GAKP01005302 JAC53650.1 AXCN02000047 CH933807 EDW11460.1 GBXI01015307 JAC98984.1 ADMH02001603 ETN61874.1 GGFJ01008802 MBW57943.1 AXCM01003145 GAMC01000929 JAC05627.1 GFDL01013166 JAV21879.1 HQ384023 ADQ64516.1 GFDG01001279 JAV17520.1 APCN01000459 GDIP01125362 JAL78352.1 GL449694 EFN82209.1 GDIP01149607 LRGB01002101 JAJ73795.1 KZS09182.1 GDIQ01088466 JAN06271.1 GBHO01034775 GBHO01034773 GBRD01000224 GDHC01018026 GDHC01004997 GDHC01001532 JAG08829.1 JAG08831.1 JAG65597.1 JAQ00603.1 JAQ13632.1 JAQ17097.1 GGFK01011224 MBW44545.1 CH963920 EDW78167.1 GGFK01011384 MBW44705.1 AAAB01008859 EAA07461.4 GFDG01001276 JAV17523.1 JRES01001272 KNC24038.1 OUUW01000004 SPP79837.1 GFDL01012835 JAV22210.1 JXUM01013055 KQ560367 KXJ82782.1 AE014134 BT030995 AAF51108.3 ABV82377.1 GAPW01003876 JAC09722.1 CH940649 EDW64785.1 ABJB010600558 ABJB011069411 DS921183 EEC17249.1 CH676463 EDW44610.1 CM000157 EDW87537.1 CM000361 CM002910 EDX03614.1 KMY87891.1 GDIP01098811 JAM04904.1 GBGD01002820 JAC86069.1 GDKW01002544 JAI54051.1 GBYB01010614 JAG80381.1 GEFM01004801 JAP70995.1 CH916368 EDW04048.1 LBMM01006587 KMQ90488.1 GADI01006805 JAA67003.1 KQ414670 KOC64291.1 CH477623 EAT38145.1 KQ977791 KYM99692.1 GANO01003443 JAB56428.1 AJWK01031750 KK120106 KFM77539.1 GECL01000156 JAP05968.1 CH954177 EDV57620.1 KQ976396 KYM93014.1 EAAA01001392 GDAI01001830 JAI15773.1 NNAY01002666 OXU20822.1 KK107364 QOIP01000007 EZA51947.1 RLU20917.1 BC122499 CM004483 AAI22500.1 OCT60986.1 CH478337 EAT33207.1
PZC77527.1 NWSH01000813 PCG74084.1 AGBW02012702 OWR44584.1 GDQN01008790 GDQN01008107 JAT82264.1 JAT82947.1 GAIX01012193 JAA80367.1 GDQN01008121 JAT82933.1 GEZM01084997 GEZM01084996 JAV60329.1 GEDC01027896 JAS09402.1 KQ971338 EFA02191.1 GECU01034735 JAS72971.1 ATLV01012143 KE524764 KFB36213.1 APGK01039000 KB740966 KB632186 ENN76930.1 ERL89700.1 GDHF01006501 JAI45813.1 GAKP01005302 JAC53650.1 AXCN02000047 CH933807 EDW11460.1 GBXI01015307 JAC98984.1 ADMH02001603 ETN61874.1 GGFJ01008802 MBW57943.1 AXCM01003145 GAMC01000929 JAC05627.1 GFDL01013166 JAV21879.1 HQ384023 ADQ64516.1 GFDG01001279 JAV17520.1 APCN01000459 GDIP01125362 JAL78352.1 GL449694 EFN82209.1 GDIP01149607 LRGB01002101 JAJ73795.1 KZS09182.1 GDIQ01088466 JAN06271.1 GBHO01034775 GBHO01034773 GBRD01000224 GDHC01018026 GDHC01004997 GDHC01001532 JAG08829.1 JAG08831.1 JAG65597.1 JAQ00603.1 JAQ13632.1 JAQ17097.1 GGFK01011224 MBW44545.1 CH963920 EDW78167.1 GGFK01011384 MBW44705.1 AAAB01008859 EAA07461.4 GFDG01001276 JAV17523.1 JRES01001272 KNC24038.1 OUUW01000004 SPP79837.1 GFDL01012835 JAV22210.1 JXUM01013055 KQ560367 KXJ82782.1 AE014134 BT030995 AAF51108.3 ABV82377.1 GAPW01003876 JAC09722.1 CH940649 EDW64785.1 ABJB010600558 ABJB011069411 DS921183 EEC17249.1 CH676463 EDW44610.1 CM000157 EDW87537.1 CM000361 CM002910 EDX03614.1 KMY87891.1 GDIP01098811 JAM04904.1 GBGD01002820 JAC86069.1 GDKW01002544 JAI54051.1 GBYB01010614 JAG80381.1 GEFM01004801 JAP70995.1 CH916368 EDW04048.1 LBMM01006587 KMQ90488.1 GADI01006805 JAA67003.1 KQ414670 KOC64291.1 CH477623 EAT38145.1 KQ977791 KYM99692.1 GANO01003443 JAB56428.1 AJWK01031750 KK120106 KFM77539.1 GECL01000156 JAP05968.1 CH954177 EDV57620.1 KQ976396 KYM93014.1 EAAA01001392 GDAI01001830 JAI15773.1 NNAY01002666 OXU20822.1 KK107364 QOIP01000007 EZA51947.1 RLU20917.1 BC122499 CM004483 AAI22500.1 OCT60986.1 CH478337 EAT33207.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000218220
UP000007151
UP000079169
+ More
UP000007266 UP000030765 UP000019118 UP000030742 UP000075886 UP000009192 UP000075900 UP000075880 UP000000673 UP000075883 UP000075920 UP000075840 UP000008237 UP000076858 UP000007798 UP000075882 UP000076407 UP000075884 UP000075903 UP000007062 UP000037069 UP000075902 UP000268350 UP000075885 UP000076408 UP000069940 UP000249989 UP000000803 UP000075881 UP000095301 UP000008792 UP000001555 UP000001292 UP000002282 UP000000304 UP000076420 UP000001070 UP000036403 UP000053825 UP000091820 UP000095300 UP000008820 UP000078542 UP000092461 UP000054359 UP000092445 UP000008711 UP000078540 UP000008144 UP000215335 UP000053097 UP000279307 UP000192221 UP000085678 UP000002358 UP000186698
UP000007266 UP000030765 UP000019118 UP000030742 UP000075886 UP000009192 UP000075900 UP000075880 UP000000673 UP000075883 UP000075920 UP000075840 UP000008237 UP000076858 UP000007798 UP000075882 UP000076407 UP000075884 UP000075903 UP000007062 UP000037069 UP000075902 UP000268350 UP000075885 UP000076408 UP000069940 UP000249989 UP000000803 UP000075881 UP000095301 UP000008792 UP000001555 UP000001292 UP000002282 UP000000304 UP000076420 UP000001070 UP000036403 UP000053825 UP000091820 UP000095300 UP000008820 UP000078542 UP000092461 UP000054359 UP000092445 UP000008711 UP000078540 UP000008144 UP000215335 UP000053097 UP000279307 UP000192221 UP000085678 UP000002358 UP000186698
SUPFAM
SSF118310
SSF118310
Gene 3D
ProteinModelPortal
H9JIV2
A0A0N0PA85
A0A437BPS7
A0A2W1BWE1
A0A2A4JR49
A0A212ET10
+ More
A0A1E1W7M5 S4PTE2 A0A1E1W7I5 A0A1Y1KFR5 A0A1B6C7F1 A0A1S3D3G5 D2A2F5 A0A1B6HEE5 A0A084VE19 N6U5N9 A0A0K8W3Q9 A0A034WHM7 A0A182QTW0 B4KJL5 A0A0A1WKC5 A0A182RNL7 A0A182INA2 W5JGC7 A0A2M4BY54 A0A182LYQ0 W8CAZ0 A0A1Q3F2X5 A0A182VQ82 E5L9P2 A0A1L8EFL7 A0A182HTC8 A0A0P5TN19 E2BQ21 A0A0P5E104 A0A0P6CQU6 A0A0A9WM26 A0A2M4AUT6 B4N105 A0A182LFH9 A0A182XEI2 A0A182N4L7 A0A2M4AVA9 A0A182VN11 Q7QBW5 A0A1L8EG21 A0A0L0BVM4 A0A182TI46 A0A3B0JBU9 A0A1Q3F3S8 A0A182PTH4 A0A182Y7B0 A0A182GJX2 Q9VQQ7 A0A182K5M3 A0A1I8MPU0 A0A023EKP1 B4LSG3 B7QEH7 B4INK1 B4NXW6 B4Q9K3 A0A2C9LW43 A0A0P5VKA4 A0A069DXJ0 A0A0P4VIZ5 A0A2C9LW62 A0A0C9R2Z5 A0A131XXQ4 B4JBU5 A0A0J7KJ86 A0A0K8R7Z4 A0A0L7R0F8 A0A1A9WXK8 A0A1I8PVE6 Q16UF2 A0A1S4G260 A0A195CG29 U5ES23 A0A1B0CW47 A0A087UJK0 A0A0V0GD02 A0A1A9Z7W1 B3N321 A0A195BWK5 F6R192 A0A0K8TNQ6 A0A232ER66 A0A026W763 A0A1W4UUH4 A0A1S3IZ84 A0A1S3IZT3 A0A1S3J062 K7J1D4 Q0P3R6 A0A1S3IZL5 Q16G40
A0A1E1W7M5 S4PTE2 A0A1E1W7I5 A0A1Y1KFR5 A0A1B6C7F1 A0A1S3D3G5 D2A2F5 A0A1B6HEE5 A0A084VE19 N6U5N9 A0A0K8W3Q9 A0A034WHM7 A0A182QTW0 B4KJL5 A0A0A1WKC5 A0A182RNL7 A0A182INA2 W5JGC7 A0A2M4BY54 A0A182LYQ0 W8CAZ0 A0A1Q3F2X5 A0A182VQ82 E5L9P2 A0A1L8EFL7 A0A182HTC8 A0A0P5TN19 E2BQ21 A0A0P5E104 A0A0P6CQU6 A0A0A9WM26 A0A2M4AUT6 B4N105 A0A182LFH9 A0A182XEI2 A0A182N4L7 A0A2M4AVA9 A0A182VN11 Q7QBW5 A0A1L8EG21 A0A0L0BVM4 A0A182TI46 A0A3B0JBU9 A0A1Q3F3S8 A0A182PTH4 A0A182Y7B0 A0A182GJX2 Q9VQQ7 A0A182K5M3 A0A1I8MPU0 A0A023EKP1 B4LSG3 B7QEH7 B4INK1 B4NXW6 B4Q9K3 A0A2C9LW43 A0A0P5VKA4 A0A069DXJ0 A0A0P4VIZ5 A0A2C9LW62 A0A0C9R2Z5 A0A131XXQ4 B4JBU5 A0A0J7KJ86 A0A0K8R7Z4 A0A0L7R0F8 A0A1A9WXK8 A0A1I8PVE6 Q16UF2 A0A1S4G260 A0A195CG29 U5ES23 A0A1B0CW47 A0A087UJK0 A0A0V0GD02 A0A1A9Z7W1 B3N321 A0A195BWK5 F6R192 A0A0K8TNQ6 A0A232ER66 A0A026W763 A0A1W4UUH4 A0A1S3IZ84 A0A1S3IZT3 A0A1S3J062 K7J1D4 Q0P3R6 A0A1S3IZL5 Q16G40
PDB
1X4V
E-value=2.59745e-07,
Score=128
Ontologies
GO
Topology
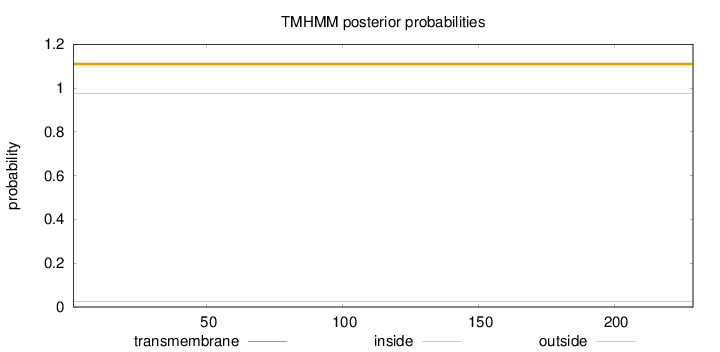
Length:
229
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02655
outside
1 - 229
Population Genetic Test Statistics
Pi
244.688745
Theta
204.691573
Tajima's D
0.91607
CLR
0.225134
CSRT
0.634068296585171
Interpretation
Uncertain
