Gene
KWMTBOMO08238 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009454
Annotation
PREDICTED:_uncharacterized_protein_C9orf78_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.487
Sequence
CDS
ATGGCAGATTCAGATTCTCAAGAAAATCGACAAGTCGAAGAAATTAAATTGAAATCTAAGAAGCCTAGAAATCTTCGTACGCGTATCAAAGATGAAGACTCTGATGATGAGGAGCACATTTTGCAAAGACTTCAAGAAACAAAAGAAATTCAGAAACTTCGAGAAAGACCTAATGGGGTGAGCATCATAGCCCTGGCAACCGGCGAGAAAGTAACGTTGGAAGATGAAATGACTTGTAAAGATCCTTTTAAAGTTAAAACTGGCGGGATGATTAACATGCAAGCCCTTAAGAACGGGAAACTGAAGCAGGTGGATGATGCGTATGATACCGGAATAGGCACACAATTTTCAGCGGAAACGAACAAACGCGACGAAGATGAGGAGATGATGAAGTACATCGAAGAACAACTTGCAAAAAGAAAAGAGGGATGTGACAAAGACAATAAAGACCACAACCATACAGAAACACTAAAGTATTTGTCTCCCGAGGAAGCAGCCTTACTTTCCTTGCCAGACCATTTGAGAGTCTCTTCAAATCAGAGGTCAGAAGAAATGCTTTCTAATCAGATGCTCAGTGGTATACCAGAAGTAGATTTAGGCATTGACGCGAAAATAAAAAACATTGAGGCAACTGAAGAGGCCAAGATGAAATTAATATGGGAAAGACAAAACAAAAAAGATGGCCCATCTCAATTTGTTCCAACAAATATGGCCGTTAACTTCGTTCAGCATAACAGATTTAACATGGAGAATGATAAAAAACGAAAAATTGAAAAAGTTGTAGTGCCCAAAACAGAAATTAGCGTGATTGATGAGAACGTTGATAAAATAGTTAAGAAAGCTAAAGGGGAAAGAGCAACTGATGATTATCATTATGAGAAGTTTAAAAAACAGTTTAGAAGGTGTTAA
Protein
MADSDSQENRQVEEIKLKSKKPRNLRTRIKDEDSDDEEHILQRLQETKEIQKLRERPNGVSIIALATGEKVTLEDEMTCKDPFKVKTGGMINMQALKNGKLKQVDDAYDTGIGTQFSAETNKRDEDEEMMKYIEEQLAKRKEGCDKDNKDHNHTETLKYLSPEEAALLSLPDHLRVSSNQRSEEMLSNQMLSGIPEVDLGIDAKIKNIEATEEAKMKLIWERQNKKDGPSQFVPTNMAVNFVQHNRFNMENDKKRKIEKVVVPKTEISVIDENVDKIVKKAKGERATDDYHYEKFKKQFRRC
Summary
Uniprot
H9JIV3
A0A3S2TFE6
A0A0N1INC4
A0A2A4JQP0
A0A0N1IQ55
A0A2W1BV67
+ More
A0FDQ9 A0A1E1W326 S4P903 A0A1E1WEV4 A0A1S3DAJ6 A0A1B6EU69 A0A2P8YUQ9 A0A1B6ISM9 A0A212ERM7 A0A2J7PW63 A0A1Q3F6F3 A0A034V1A0 A0A1B6M0Q1 A0A1L8DQW4 D6WEK7 A0A2S2QFL6 J3JX03 A0A067QR53 U5EZG7 U4UKD0 Q16RG0 B0WAM6 A0A0A1WSZ6 B4MND3 W8CCA8 B3M7R5 B4L969 A0A182GWV7 A0A0L7QXT7 B4LBU5 B4PDR6 A0A0P4VMD5 R4G3R5 A0A069DYI6 A0A0Q9XUV8 A0A2H8TTL6 A0A0L0C0E3 B4J1X9 B3NET2 A0A224XUP7 A0A0J9RLY1 B4HVX8 A0A2M3ZBC2 A0A2M3ZIW0 Q9W0A9 C4WWZ4 B4K3K0 A0A2M4BV40 A0A2M4BVA8 A0A182QAR3 A0A1W4W6E1 A0A182PMV9 A0A1I8NHN4 A0A1I8NQ27 A0A1B0BQZ5 A0A2M4AX34 A0A182NI86 A0A1A9XQJ1 A0A023F8H9 A0A182FGF0 A0A0C9QIM4 A0A146L667 A0A0A9ZG77 E9FWZ6 A0A0V0G9Q5 A0A1A9ZTQ6 A0A182J6U0 A0A1A9V4J3 A0A026WDV8 A0A182WBP6 A0A1I8JUH6 W5J863 A0A0T6BAP8 A0A3B0JW70 A0A1D2N497 A0A2A3EMW5 A0A182JU24 A0A084WGK4 Q2LZP5 B4H3F7 Q7Q562 A0A1W4WN24 K7J6B7 A0A182TQU1 A0A182V676 A0A1B0GAX0 A0A336LWJ6 A0A158ND56 A0A1A9WPX5 A0A0K8U7E6 F4X7C0 A0A182X4K3 A0A195FHV3 A0A0P6FS71
A0FDQ9 A0A1E1W326 S4P903 A0A1E1WEV4 A0A1S3DAJ6 A0A1B6EU69 A0A2P8YUQ9 A0A1B6ISM9 A0A212ERM7 A0A2J7PW63 A0A1Q3F6F3 A0A034V1A0 A0A1B6M0Q1 A0A1L8DQW4 D6WEK7 A0A2S2QFL6 J3JX03 A0A067QR53 U5EZG7 U4UKD0 Q16RG0 B0WAM6 A0A0A1WSZ6 B4MND3 W8CCA8 B3M7R5 B4L969 A0A182GWV7 A0A0L7QXT7 B4LBU5 B4PDR6 A0A0P4VMD5 R4G3R5 A0A069DYI6 A0A0Q9XUV8 A0A2H8TTL6 A0A0L0C0E3 B4J1X9 B3NET2 A0A224XUP7 A0A0J9RLY1 B4HVX8 A0A2M3ZBC2 A0A2M3ZIW0 Q9W0A9 C4WWZ4 B4K3K0 A0A2M4BV40 A0A2M4BVA8 A0A182QAR3 A0A1W4W6E1 A0A182PMV9 A0A1I8NHN4 A0A1I8NQ27 A0A1B0BQZ5 A0A2M4AX34 A0A182NI86 A0A1A9XQJ1 A0A023F8H9 A0A182FGF0 A0A0C9QIM4 A0A146L667 A0A0A9ZG77 E9FWZ6 A0A0V0G9Q5 A0A1A9ZTQ6 A0A182J6U0 A0A1A9V4J3 A0A026WDV8 A0A182WBP6 A0A1I8JUH6 W5J863 A0A0T6BAP8 A0A3B0JW70 A0A1D2N497 A0A2A3EMW5 A0A182JU24 A0A084WGK4 Q2LZP5 B4H3F7 Q7Q562 A0A1W4WN24 K7J6B7 A0A182TQU1 A0A182V676 A0A1B0GAX0 A0A336LWJ6 A0A158ND56 A0A1A9WPX5 A0A0K8U7E6 F4X7C0 A0A182X4K3 A0A195FHV3 A0A0P6FS71
Pubmed
19121390
26354079
28756777
23622113
29403074
22118469
+ More
25348373 18362917 19820115 22516182 23537049 24845553 17510324 25830018 17994087 24495485 26483478 17550304 27129103 26334808 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25474469 26823975 25401762 21292972 24508170 30249741 20920257 23761445 27289101 24438588 15632085 12364791 14747013 17210077 20075255 21347285 21719571
25348373 18362917 19820115 22516182 23537049 24845553 17510324 25830018 17994087 24495485 26483478 17550304 27129103 26334808 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25474469 26823975 25401762 21292972 24508170 30249741 20920257 23761445 27289101 24438588 15632085 12364791 14747013 17210077 20075255 21347285 21719571
EMBL
BABH01023137
RSAL01000196
RVE44547.1
KQ458751
KPJ05322.1
NWSH01000813
+ More
PCG74086.1 KQ459986 KPJ18854.1 KZ149927 PZC77524.1 DQ985600 ABJ97189.1 GDQN01009679 JAT81375.1 GAIX01007037 JAA85523.1 GDQN01005673 JAT85381.1 GECZ01028267 JAS41502.1 PYGN01000349 PSN47991.1 GECU01017772 JAS89934.1 AGBW02012961 OWR44156.1 NEVH01020938 PNF20569.1 GFDL01011900 JAV23145.1 GAKP01022683 JAC36269.1 GEBQ01010480 JAT29497.1 GFDF01005218 JAV08866.1 KQ971318 EFA00334.1 GGMS01007332 MBY76535.1 APGK01043942 BT127771 KB741019 AEE62733.1 ENN75146.1 KK853058 KDR11927.1 GANO01000829 JAB59042.1 KB632338 ERL92923.1 CH477712 EAT36987.1 DS231873 EDS41580.1 GBXI01012749 JAD01543.1 CH963847 EDW72642.1 GAMC01004021 JAC02535.1 CH902618 EDV38788.1 CH933816 EDW17244.1 JXUM01094229 KQ564113 KXJ72765.1 KQ414699 KOC63432.1 CH940647 EDW68722.1 CM000159 EDW92881.1 GDKW01000292 JAI56303.1 ACPB03000182 GAHY01001990 JAA75520.1 GBGD01002325 JAC86564.1 KRG07688.1 GFXV01005790 MBW17595.1 JRES01001077 KNC25721.1 CH916366 EDV95904.1 CH954178 EDV50205.1 GFTR01004755 JAW11671.1 CM002912 KMY96918.1 CH480817 EDW50093.1 GGFM01004997 MBW25748.1 GGFM01007733 MBW28484.1 AE014296 AY069253 AAF47544.1 AAL39398.1 ABLF02029284 AK342359 BAH72414.1 CH923264 EDW04423.1 GGFJ01007794 MBW56935.1 GGFJ01007793 MBW56934.1 AXCN02000877 JXJN01018858 GGFK01012038 MBW45359.1 GBBI01001085 JAC17627.1 GBYB01014598 JAG84365.1 GDHC01016459 JAQ02170.1 GBHO01002639 GBRD01012420 GDHC01018202 JAG40965.1 JAG53404.1 JAQ00427.1 GL732526 EFX87982.1 GECL01001620 JAP04504.1 KK107260 QOIP01000001 EZA54252.1 RLU26244.1 ADMH02002104 ETN59055.1 LJIG01002567 KRT84373.1 OUUW01000002 SPP76961.1 LJIJ01000234 ODN00093.1 KZ288212 PBC32834.1 ATLV01023588 KE525344 KFB49348.1 CH379069 EAL29462.1 CH479206 EDW30897.1 AAAB01008960 EAA11347.2 CCAG010001198 UFQT01000253 SSX22426.1 ADTU01012403 GDHF01029720 JAI22594.1 GL888828 EGI57732.1 KQ981560 KYN39847.1 GDIQ01054527 LRGB01001564 JAN40210.1 KZS11799.1
PCG74086.1 KQ459986 KPJ18854.1 KZ149927 PZC77524.1 DQ985600 ABJ97189.1 GDQN01009679 JAT81375.1 GAIX01007037 JAA85523.1 GDQN01005673 JAT85381.1 GECZ01028267 JAS41502.1 PYGN01000349 PSN47991.1 GECU01017772 JAS89934.1 AGBW02012961 OWR44156.1 NEVH01020938 PNF20569.1 GFDL01011900 JAV23145.1 GAKP01022683 JAC36269.1 GEBQ01010480 JAT29497.1 GFDF01005218 JAV08866.1 KQ971318 EFA00334.1 GGMS01007332 MBY76535.1 APGK01043942 BT127771 KB741019 AEE62733.1 ENN75146.1 KK853058 KDR11927.1 GANO01000829 JAB59042.1 KB632338 ERL92923.1 CH477712 EAT36987.1 DS231873 EDS41580.1 GBXI01012749 JAD01543.1 CH963847 EDW72642.1 GAMC01004021 JAC02535.1 CH902618 EDV38788.1 CH933816 EDW17244.1 JXUM01094229 KQ564113 KXJ72765.1 KQ414699 KOC63432.1 CH940647 EDW68722.1 CM000159 EDW92881.1 GDKW01000292 JAI56303.1 ACPB03000182 GAHY01001990 JAA75520.1 GBGD01002325 JAC86564.1 KRG07688.1 GFXV01005790 MBW17595.1 JRES01001077 KNC25721.1 CH916366 EDV95904.1 CH954178 EDV50205.1 GFTR01004755 JAW11671.1 CM002912 KMY96918.1 CH480817 EDW50093.1 GGFM01004997 MBW25748.1 GGFM01007733 MBW28484.1 AE014296 AY069253 AAF47544.1 AAL39398.1 ABLF02029284 AK342359 BAH72414.1 CH923264 EDW04423.1 GGFJ01007794 MBW56935.1 GGFJ01007793 MBW56934.1 AXCN02000877 JXJN01018858 GGFK01012038 MBW45359.1 GBBI01001085 JAC17627.1 GBYB01014598 JAG84365.1 GDHC01016459 JAQ02170.1 GBHO01002639 GBRD01012420 GDHC01018202 JAG40965.1 JAG53404.1 JAQ00427.1 GL732526 EFX87982.1 GECL01001620 JAP04504.1 KK107260 QOIP01000001 EZA54252.1 RLU26244.1 ADMH02002104 ETN59055.1 LJIG01002567 KRT84373.1 OUUW01000002 SPP76961.1 LJIJ01000234 ODN00093.1 KZ288212 PBC32834.1 ATLV01023588 KE525344 KFB49348.1 CH379069 EAL29462.1 CH479206 EDW30897.1 AAAB01008960 EAA11347.2 CCAG010001198 UFQT01000253 SSX22426.1 ADTU01012403 GDHF01029720 JAI22594.1 GL888828 EGI57732.1 KQ981560 KYN39847.1 GDIQ01054527 LRGB01001564 JAN40210.1 KZS11799.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000053240
UP000079169
+ More
UP000245037 UP000007151 UP000235965 UP000007266 UP000019118 UP000027135 UP000030742 UP000008820 UP000002320 UP000007798 UP000007801 UP000009192 UP000069940 UP000249989 UP000053825 UP000008792 UP000002282 UP000015103 UP000037069 UP000001070 UP000008711 UP000001292 UP000000803 UP000007819 UP000075886 UP000192221 UP000075885 UP000095301 UP000095300 UP000092460 UP000075884 UP000092443 UP000069272 UP000000305 UP000092445 UP000075880 UP000078200 UP000053097 UP000279307 UP000075920 UP000075900 UP000000673 UP000268350 UP000094527 UP000242457 UP000075881 UP000030765 UP000001819 UP000008744 UP000007062 UP000192223 UP000002358 UP000075902 UP000075903 UP000092444 UP000005205 UP000091820 UP000007755 UP000076407 UP000078541 UP000076858
UP000245037 UP000007151 UP000235965 UP000007266 UP000019118 UP000027135 UP000030742 UP000008820 UP000002320 UP000007798 UP000007801 UP000009192 UP000069940 UP000249989 UP000053825 UP000008792 UP000002282 UP000015103 UP000037069 UP000001070 UP000008711 UP000001292 UP000000803 UP000007819 UP000075886 UP000192221 UP000075885 UP000095301 UP000095300 UP000092460 UP000075884 UP000092443 UP000069272 UP000000305 UP000092445 UP000075880 UP000078200 UP000053097 UP000279307 UP000075920 UP000075900 UP000000673 UP000268350 UP000094527 UP000242457 UP000075881 UP000030765 UP000001819 UP000008744 UP000007062 UP000192223 UP000002358 UP000075902 UP000075903 UP000092444 UP000005205 UP000091820 UP000007755 UP000076407 UP000078541 UP000076858
Pfam
PF07052 Hep_59
Interpro
IPR010756
Tls1
ProteinModelPortal
H9JIV3
A0A3S2TFE6
A0A0N1INC4
A0A2A4JQP0
A0A0N1IQ55
A0A2W1BV67
+ More
A0FDQ9 A0A1E1W326 S4P903 A0A1E1WEV4 A0A1S3DAJ6 A0A1B6EU69 A0A2P8YUQ9 A0A1B6ISM9 A0A212ERM7 A0A2J7PW63 A0A1Q3F6F3 A0A034V1A0 A0A1B6M0Q1 A0A1L8DQW4 D6WEK7 A0A2S2QFL6 J3JX03 A0A067QR53 U5EZG7 U4UKD0 Q16RG0 B0WAM6 A0A0A1WSZ6 B4MND3 W8CCA8 B3M7R5 B4L969 A0A182GWV7 A0A0L7QXT7 B4LBU5 B4PDR6 A0A0P4VMD5 R4G3R5 A0A069DYI6 A0A0Q9XUV8 A0A2H8TTL6 A0A0L0C0E3 B4J1X9 B3NET2 A0A224XUP7 A0A0J9RLY1 B4HVX8 A0A2M3ZBC2 A0A2M3ZIW0 Q9W0A9 C4WWZ4 B4K3K0 A0A2M4BV40 A0A2M4BVA8 A0A182QAR3 A0A1W4W6E1 A0A182PMV9 A0A1I8NHN4 A0A1I8NQ27 A0A1B0BQZ5 A0A2M4AX34 A0A182NI86 A0A1A9XQJ1 A0A023F8H9 A0A182FGF0 A0A0C9QIM4 A0A146L667 A0A0A9ZG77 E9FWZ6 A0A0V0G9Q5 A0A1A9ZTQ6 A0A182J6U0 A0A1A9V4J3 A0A026WDV8 A0A182WBP6 A0A1I8JUH6 W5J863 A0A0T6BAP8 A0A3B0JW70 A0A1D2N497 A0A2A3EMW5 A0A182JU24 A0A084WGK4 Q2LZP5 B4H3F7 Q7Q562 A0A1W4WN24 K7J6B7 A0A182TQU1 A0A182V676 A0A1B0GAX0 A0A336LWJ6 A0A158ND56 A0A1A9WPX5 A0A0K8U7E6 F4X7C0 A0A182X4K3 A0A195FHV3 A0A0P6FS71
A0FDQ9 A0A1E1W326 S4P903 A0A1E1WEV4 A0A1S3DAJ6 A0A1B6EU69 A0A2P8YUQ9 A0A1B6ISM9 A0A212ERM7 A0A2J7PW63 A0A1Q3F6F3 A0A034V1A0 A0A1B6M0Q1 A0A1L8DQW4 D6WEK7 A0A2S2QFL6 J3JX03 A0A067QR53 U5EZG7 U4UKD0 Q16RG0 B0WAM6 A0A0A1WSZ6 B4MND3 W8CCA8 B3M7R5 B4L969 A0A182GWV7 A0A0L7QXT7 B4LBU5 B4PDR6 A0A0P4VMD5 R4G3R5 A0A069DYI6 A0A0Q9XUV8 A0A2H8TTL6 A0A0L0C0E3 B4J1X9 B3NET2 A0A224XUP7 A0A0J9RLY1 B4HVX8 A0A2M3ZBC2 A0A2M3ZIW0 Q9W0A9 C4WWZ4 B4K3K0 A0A2M4BV40 A0A2M4BVA8 A0A182QAR3 A0A1W4W6E1 A0A182PMV9 A0A1I8NHN4 A0A1I8NQ27 A0A1B0BQZ5 A0A2M4AX34 A0A182NI86 A0A1A9XQJ1 A0A023F8H9 A0A182FGF0 A0A0C9QIM4 A0A146L667 A0A0A9ZG77 E9FWZ6 A0A0V0G9Q5 A0A1A9ZTQ6 A0A182J6U0 A0A1A9V4J3 A0A026WDV8 A0A182WBP6 A0A1I8JUH6 W5J863 A0A0T6BAP8 A0A3B0JW70 A0A1D2N497 A0A2A3EMW5 A0A182JU24 A0A084WGK4 Q2LZP5 B4H3F7 Q7Q562 A0A1W4WN24 K7J6B7 A0A182TQU1 A0A182V676 A0A1B0GAX0 A0A336LWJ6 A0A158ND56 A0A1A9WPX5 A0A0K8U7E6 F4X7C0 A0A182X4K3 A0A195FHV3 A0A0P6FS71
Ontologies
PANTHER
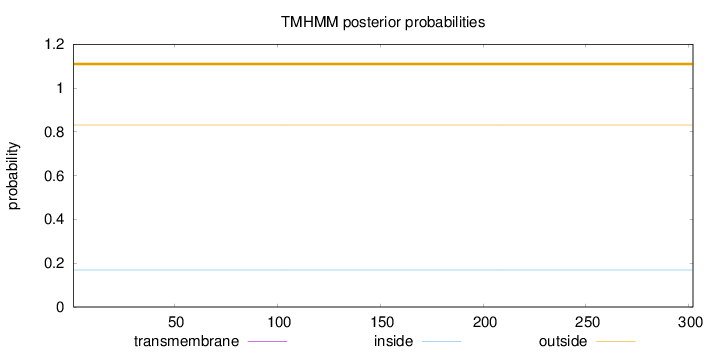
Topology
Length:
302
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00086
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.16880
outside
1 - 302
Population Genetic Test Statistics
Pi
177.938475
Theta
133.746795
Tajima's D
-0.700017
CLR
1048.945305
CSRT
0.198090095495225
Interpretation
Uncertain
