Gene
KWMTBOMO08228
Pre Gene Modal
BGIBMGA014572
Annotation
PREDICTED:_GDP-Man:Man(3)GlcNAc(2)-PP-Dol_alpha-1?2-mannosyltransferase_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 1.486 PlasmaMembrane Reliability : 1.697
Sequence
CDS
ATGAGAATTAAAGAACGGAAACTGGAGAGATTGAAACAACATGGACCAAACGTGGCATTCTTCCATCCCTACTGTAATGCTGGAGGTGGGGGTGAAAGAGTACTGTGGGTGGCCATCAAAGCTGTCCTAACCAGGTATCCAGATTCAAATATTTTCATCTATACAGTGGAAACAGAAGAACCTCAAAAAATACTAGATAAAGCACAGAATCACTTCAATGTGAAATTGGACTTGAAGAAGATAACATTTGTACATCTTACTTTGAAAATCTTGATTGAAGCAAAAACATACCCATACTTTACCTTAGTGCTCCAAAGCCTTGGATCTATGGTGCTCGGAATGGAAGCATTTTTGAAACTAAATCCTGATGTTTACATAGACACGATGGGATGTGCCTTCACGTTCCCAATATTCCGTTATTTGGCTCAGTCCAGGGTCGGATGTTACGTCCATTATCCCACCATAACGACAGCAATGATGCGCAGGGTCAAACACAGGATAGTTTCATACAACAATTCTGGTATAATTGCCAGGAATCCTTTGTACACTTGGATAAAGTTGCTGTATTATAAGATATTTGGTTGGTTATACGGTATTGTGGGTCGGTGTGCAGACGTTATAATGGTAAATGGAACTTGGACAGAGGAACATATCAATGAACTTTGGAAGGAACCTTACAACACTTACAAAGTCTTCCCACCTTGTGAGGTTACAGAATTGAAACAATTACGTTCTTTAGTGAAAGAGTCAGATCCTATCCGGATCCTGTCGGTCGCCCAGTTCAGACCGGAGAAAGATCATCCGCTGATGCTGCAGGCGATGTACGAGCTGAGGAGCCTGTTGACCAAGAATGAATTCCTTTGGAATAAGATCCAGTTGGTTCTGGCGGGTTCCTGCCGCAATGCCGAAGACGAGGAGCGCGTGAGACACCTCAAGGACCTGGCCAAGCACTTGTCCTTAGAAAACAACGTGCAGTTTGAGGTGAACGTGACGTACGCTCGACTCTTGCACCTTTACCAGACCAGCCTCATCGGTCTTCACGCCATGTGGAATGAGCACTTCGGAATCAGCGTGGTGGAGAGCATGGCGGCGGGGCTGGTGACGGTGGCGCACCGCTCGGGCGGGCCGCGCGCCGACATCGTGGCGCCGCACGCCGGCTACCTGGCCGCCGAGCCCGAGCAGTACGCCGCCGCGCTGCTGCACGCGCTGTCGCTGTCCGCCGCCAGCCGCGCCGACCTCGTGGCCGCCGCGCGTGCATCCGTGGATCGTTTCTCGGTGAAGGAGTTTGAGAAATCATTCCTTCGAGCGTGCGAACCGTTAATGTCTTTCTAG
Protein
MRIKERKLERLKQHGPNVAFFHPYCNAGGGGERVLWVAIKAVLTRYPDSNIFIYTVETEEPQKILDKAQNHFNVKLDLKKITFVHLTLKILIEAKTYPYFTLVLQSLGSMVLGMEAFLKLNPDVYIDTMGCAFTFPIFRYLAQSRVGCYVHYPTITTAMMRRVKHRIVSYNNSGIIARNPLYTWIKLLYYKIFGWLYGIVGRCADVIMVNGTWTEEHINELWKEPYNTYKVFPPCEVTELKQLRSLVKESDPIRILSVAQFRPEKDHPLMLQAMYELRSLLTKNEFLWNKIQLVLAGSCRNAEDEERVRHLKDLAKHLSLENNVQFEVNVTYARLLHLYQTSLIGLHAMWNEHFGISVVESMAAGLVTVAHRSGGPRADIVAPHAGYLAAEPEQYAAALLHALSLSAASRADLVAAARASVDRFSVKEFEKSFLRACEPLMSF
Summary
Uniprot
A0A2H1VRD4
A0A194PZ26
A0A2W1BF51
A0A212EIK5
Q17NY5
A0A023EVB4
+ More
A0A182GVY6 A0A1S4EW70 A0A1Q3FFG0 B0XCW3 A0A182N3D4 A0A182M4R0 Q7QHR9 A0A1S4H6C1 A0A182HY59 A0A182LHA7 A0A182WXI5 A0A182Q1T1 A0A182UQ95 A0A182RLK3 A0A182TM15 A0A182P2M8 A0A182IWX4 A0A182SD74 A0A182Y6Z5 A0A182WFL6 U5EYS0 B4PDJ4 B4ITE5 B4IAT2 A0A0J9S0X7 B3NEC2 A0A084VR10 A0A0K8TTR7 A0A336JYF7 A0A0P8XVR5 Q9VP06 A0A2M4BLK1 A0A1L8E3H8 A0A1L8E440 A0A1W4UT35 W5JRN5 B4MXP7 A0A0A1WVP7 A0A182FLH6 A0A034WRX4 W8BI12 A0A3B0JZ56 A0A2M3Z6C8 A0A2M4ACP7 Q29F06 A0A1I8P6Q8 A0A0Q9XE21 A0A1J1HP19 B4KXR0 D3TQ23 A0A1A9ZFZ1 T1H6F0 A0A1A9WTJ2 A0A1I8N3J9 A0A1A9V302 A0A1A9YL22 A0A1B0AR97 B4J1S3 E2BDI4 A0A0L0C6G6 E2A8F9 A0A195D4H0 A0A158NHF4 F4WD88 A0A195AZY6 A0A151XFD8 A0A067QZY2 E0VLJ1 A0A2J7RI51 A0A195FW16 A0A154PBT7 A0A195E231 K7IZ54 A0A088ARN8 A0A0L7RD18 B4LFP1 A0A310SI54 A0A2A3ELG3 A0A232EUD3 A0A0C9RQW4 A0A3L8DHD7 A0A1S3CU04 A0A0N0U4U7 A0A1W4WXL8 A0A2P8YLW5 A0A224XQJ9 R4G308 D2A5X1 A0A0A9ZGB8 A0A146KUB5 A0A0P6H904 A0A0P5ZSJ2
A0A182GVY6 A0A1S4EW70 A0A1Q3FFG0 B0XCW3 A0A182N3D4 A0A182M4R0 Q7QHR9 A0A1S4H6C1 A0A182HY59 A0A182LHA7 A0A182WXI5 A0A182Q1T1 A0A182UQ95 A0A182RLK3 A0A182TM15 A0A182P2M8 A0A182IWX4 A0A182SD74 A0A182Y6Z5 A0A182WFL6 U5EYS0 B4PDJ4 B4ITE5 B4IAT2 A0A0J9S0X7 B3NEC2 A0A084VR10 A0A0K8TTR7 A0A336JYF7 A0A0P8XVR5 Q9VP06 A0A2M4BLK1 A0A1L8E3H8 A0A1L8E440 A0A1W4UT35 W5JRN5 B4MXP7 A0A0A1WVP7 A0A182FLH6 A0A034WRX4 W8BI12 A0A3B0JZ56 A0A2M3Z6C8 A0A2M4ACP7 Q29F06 A0A1I8P6Q8 A0A0Q9XE21 A0A1J1HP19 B4KXR0 D3TQ23 A0A1A9ZFZ1 T1H6F0 A0A1A9WTJ2 A0A1I8N3J9 A0A1A9V302 A0A1A9YL22 A0A1B0AR97 B4J1S3 E2BDI4 A0A0L0C6G6 E2A8F9 A0A195D4H0 A0A158NHF4 F4WD88 A0A195AZY6 A0A151XFD8 A0A067QZY2 E0VLJ1 A0A2J7RI51 A0A195FW16 A0A154PBT7 A0A195E231 K7IZ54 A0A088ARN8 A0A0L7RD18 B4LFP1 A0A310SI54 A0A2A3ELG3 A0A232EUD3 A0A0C9RQW4 A0A3L8DHD7 A0A1S3CU04 A0A0N0U4U7 A0A1W4WXL8 A0A2P8YLW5 A0A224XQJ9 R4G308 D2A5X1 A0A0A9ZGB8 A0A146KUB5 A0A0P6H904 A0A0P5ZSJ2
Pubmed
26354079
28756777
22118469
17510324
24945155
26483478
+ More
12364791 20966253 25244985 17994087 17550304 22936249 24438588 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25830018 25348373 24495485 15632085 20353571 25315136 20798317 26108605 21347285 21719571 24845553 20566863 20075255 28648823 30249741 29403074 18362917 19820115 25401762 26823975
12364791 20966253 25244985 17994087 17550304 22936249 24438588 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25830018 25348373 24495485 15632085 20353571 25315136 20798317 26108605 21347285 21719571 24845553 20566863 20075255 28648823 30249741 29403074 18362917 19820115 25401762 26823975
EMBL
ODYU01003815
SOQ43042.1
KQ459585
KPI98293.1
KZ150088
PZC73739.1
+ More
AGBW02014627 OWR41308.1 CH477195 EAT48413.1 GAPW01001174 JAC12424.1 JXUM01092006 JXUM01092007 JXUM01092008 JXUM01092009 KQ563958 KXJ73015.1 GFDL01008765 JAV26280.1 DS232723 EDS45158.1 AXCM01004177 AAAB01008816 EAA05076.3 APCN01004494 AXCN02001111 GANO01001852 JAB58019.1 CM000159 EDW94993.2 CH891692 EDW99658.2 CH480826 EDW44395.1 CM002912 KMZ01060.1 CH954178 EDV52686.1 ATLV01015404 KE525013 KFB40404.1 GDAI01000067 JAI17536.1 UFQS01000020 UFQT01000020 SSW97489.1 SSX17875.1 CH902618 KPU78794.1 AE014296 AY061012 AY113436 AAF51756.2 AAL28560.1 AAM29441.1 GGFJ01004818 MBW53959.1 GFDF01000791 JAV13293.1 GFDF01000779 JAV13305.1 ADMH02000374 ETN66796.1 CH963876 EDW76816.2 GBXI01011799 JAD02493.1 GAKP01002012 JAC56940.1 GAMC01005640 JAC00916.1 OUUW01000012 SPP87365.1 GGFM01003303 MBW24054.1 GGFK01005244 MBW38565.1 CH379070 EAL29903.3 CH933809 KRG06812.1 CVRI01000006 CRK87969.1 EDW19767.2 CCAG010023454 EZ423525 ADD19801.1 JXJN01002295 CH916366 EDV98003.1 GL447645 EFN86236.1 JRES01000833 KNC27988.1 GL437593 EFN70279.1 KQ976870 KYN07783.1 ADTU01015732 GL888086 EGI67815.1 KQ976694 KYM77605.1 KQ982194 KYQ59096.1 KK852809 KDR16005.1 DS235271 EEB14247.1 NEVH01003505 PNF40509.1 KQ981208 KYN44850.1 KQ434856 KZC08728.1 KQ979763 KYN19205.1 KQ414614 KOC68857.1 CH940647 EDW70359.1 KQ760869 OAD58763.1 KZ288222 PBC32112.1 NNAY01002138 OXU21962.1 GBYB01004589 GBYB01009655 GBYB01009656 JAG74356.1 JAG79422.1 JAG79423.1 QOIP01000008 RLU19736.1 KQ435814 KOX72671.1 PYGN01000502 PSN45247.1 GFTR01006177 JAW10249.1 ACPB03004983 GAHY01002111 JAA75399.1 KQ971345 EFA05431.1 GBHO01001051 GBRD01006489 JAG42553.1 JAG59332.1 GDHC01019882 GDHC01001668 JAP98746.1 JAQ16961.1 GDIP01137541 GDIP01038932 GDIQ01034961 LRGB01000915 JAL66173.1 JAN59776.1 KZS15184.1 GDIP01040060 JAM63655.1
AGBW02014627 OWR41308.1 CH477195 EAT48413.1 GAPW01001174 JAC12424.1 JXUM01092006 JXUM01092007 JXUM01092008 JXUM01092009 KQ563958 KXJ73015.1 GFDL01008765 JAV26280.1 DS232723 EDS45158.1 AXCM01004177 AAAB01008816 EAA05076.3 APCN01004494 AXCN02001111 GANO01001852 JAB58019.1 CM000159 EDW94993.2 CH891692 EDW99658.2 CH480826 EDW44395.1 CM002912 KMZ01060.1 CH954178 EDV52686.1 ATLV01015404 KE525013 KFB40404.1 GDAI01000067 JAI17536.1 UFQS01000020 UFQT01000020 SSW97489.1 SSX17875.1 CH902618 KPU78794.1 AE014296 AY061012 AY113436 AAF51756.2 AAL28560.1 AAM29441.1 GGFJ01004818 MBW53959.1 GFDF01000791 JAV13293.1 GFDF01000779 JAV13305.1 ADMH02000374 ETN66796.1 CH963876 EDW76816.2 GBXI01011799 JAD02493.1 GAKP01002012 JAC56940.1 GAMC01005640 JAC00916.1 OUUW01000012 SPP87365.1 GGFM01003303 MBW24054.1 GGFK01005244 MBW38565.1 CH379070 EAL29903.3 CH933809 KRG06812.1 CVRI01000006 CRK87969.1 EDW19767.2 CCAG010023454 EZ423525 ADD19801.1 JXJN01002295 CH916366 EDV98003.1 GL447645 EFN86236.1 JRES01000833 KNC27988.1 GL437593 EFN70279.1 KQ976870 KYN07783.1 ADTU01015732 GL888086 EGI67815.1 KQ976694 KYM77605.1 KQ982194 KYQ59096.1 KK852809 KDR16005.1 DS235271 EEB14247.1 NEVH01003505 PNF40509.1 KQ981208 KYN44850.1 KQ434856 KZC08728.1 KQ979763 KYN19205.1 KQ414614 KOC68857.1 CH940647 EDW70359.1 KQ760869 OAD58763.1 KZ288222 PBC32112.1 NNAY01002138 OXU21962.1 GBYB01004589 GBYB01009655 GBYB01009656 JAG74356.1 JAG79422.1 JAG79423.1 QOIP01000008 RLU19736.1 KQ435814 KOX72671.1 PYGN01000502 PSN45247.1 GFTR01006177 JAW10249.1 ACPB03004983 GAHY01002111 JAA75399.1 KQ971345 EFA05431.1 GBHO01001051 GBRD01006489 JAG42553.1 JAG59332.1 GDHC01019882 GDHC01001668 JAP98746.1 JAQ16961.1 GDIP01137541 GDIP01038932 GDIQ01034961 LRGB01000915 JAL66173.1 JAN59776.1 KZS15184.1 GDIP01040060 JAM63655.1
Proteomes
UP000053268
UP000007151
UP000008820
UP000069940
UP000249989
UP000002320
+ More
UP000075884 UP000075883 UP000007062 UP000075840 UP000075882 UP000076407 UP000075886 UP000075903 UP000075900 UP000075902 UP000075885 UP000075880 UP000075901 UP000076408 UP000075920 UP000002282 UP000001292 UP000008711 UP000030765 UP000007801 UP000000803 UP000192221 UP000000673 UP000007798 UP000069272 UP000268350 UP000001819 UP000095300 UP000009192 UP000183832 UP000092444 UP000092445 UP000015102 UP000091820 UP000095301 UP000078200 UP000092443 UP000092460 UP000001070 UP000008237 UP000037069 UP000000311 UP000078542 UP000005205 UP000007755 UP000078540 UP000075809 UP000027135 UP000009046 UP000235965 UP000078541 UP000076502 UP000078492 UP000002358 UP000005203 UP000053825 UP000008792 UP000242457 UP000215335 UP000279307 UP000079169 UP000053105 UP000192223 UP000245037 UP000015103 UP000007266 UP000076858
UP000075884 UP000075883 UP000007062 UP000075840 UP000075882 UP000076407 UP000075886 UP000075903 UP000075900 UP000075902 UP000075885 UP000075880 UP000075901 UP000076408 UP000075920 UP000002282 UP000001292 UP000008711 UP000030765 UP000007801 UP000000803 UP000192221 UP000000673 UP000007798 UP000069272 UP000268350 UP000001819 UP000095300 UP000009192 UP000183832 UP000092444 UP000092445 UP000015102 UP000091820 UP000095301 UP000078200 UP000092443 UP000092460 UP000001070 UP000008237 UP000037069 UP000000311 UP000078542 UP000005205 UP000007755 UP000078540 UP000075809 UP000027135 UP000009046 UP000235965 UP000078541 UP000076502 UP000078492 UP000002358 UP000005203 UP000053825 UP000008792 UP000242457 UP000215335 UP000279307 UP000079169 UP000053105 UP000192223 UP000245037 UP000015103 UP000007266 UP000076858
PRIDE
ProteinModelPortal
A0A2H1VRD4
A0A194PZ26
A0A2W1BF51
A0A212EIK5
Q17NY5
A0A023EVB4
+ More
A0A182GVY6 A0A1S4EW70 A0A1Q3FFG0 B0XCW3 A0A182N3D4 A0A182M4R0 Q7QHR9 A0A1S4H6C1 A0A182HY59 A0A182LHA7 A0A182WXI5 A0A182Q1T1 A0A182UQ95 A0A182RLK3 A0A182TM15 A0A182P2M8 A0A182IWX4 A0A182SD74 A0A182Y6Z5 A0A182WFL6 U5EYS0 B4PDJ4 B4ITE5 B4IAT2 A0A0J9S0X7 B3NEC2 A0A084VR10 A0A0K8TTR7 A0A336JYF7 A0A0P8XVR5 Q9VP06 A0A2M4BLK1 A0A1L8E3H8 A0A1L8E440 A0A1W4UT35 W5JRN5 B4MXP7 A0A0A1WVP7 A0A182FLH6 A0A034WRX4 W8BI12 A0A3B0JZ56 A0A2M3Z6C8 A0A2M4ACP7 Q29F06 A0A1I8P6Q8 A0A0Q9XE21 A0A1J1HP19 B4KXR0 D3TQ23 A0A1A9ZFZ1 T1H6F0 A0A1A9WTJ2 A0A1I8N3J9 A0A1A9V302 A0A1A9YL22 A0A1B0AR97 B4J1S3 E2BDI4 A0A0L0C6G6 E2A8F9 A0A195D4H0 A0A158NHF4 F4WD88 A0A195AZY6 A0A151XFD8 A0A067QZY2 E0VLJ1 A0A2J7RI51 A0A195FW16 A0A154PBT7 A0A195E231 K7IZ54 A0A088ARN8 A0A0L7RD18 B4LFP1 A0A310SI54 A0A2A3ELG3 A0A232EUD3 A0A0C9RQW4 A0A3L8DHD7 A0A1S3CU04 A0A0N0U4U7 A0A1W4WXL8 A0A2P8YLW5 A0A224XQJ9 R4G308 D2A5X1 A0A0A9ZGB8 A0A146KUB5 A0A0P6H904 A0A0P5ZSJ2
A0A182GVY6 A0A1S4EW70 A0A1Q3FFG0 B0XCW3 A0A182N3D4 A0A182M4R0 Q7QHR9 A0A1S4H6C1 A0A182HY59 A0A182LHA7 A0A182WXI5 A0A182Q1T1 A0A182UQ95 A0A182RLK3 A0A182TM15 A0A182P2M8 A0A182IWX4 A0A182SD74 A0A182Y6Z5 A0A182WFL6 U5EYS0 B4PDJ4 B4ITE5 B4IAT2 A0A0J9S0X7 B3NEC2 A0A084VR10 A0A0K8TTR7 A0A336JYF7 A0A0P8XVR5 Q9VP06 A0A2M4BLK1 A0A1L8E3H8 A0A1L8E440 A0A1W4UT35 W5JRN5 B4MXP7 A0A0A1WVP7 A0A182FLH6 A0A034WRX4 W8BI12 A0A3B0JZ56 A0A2M3Z6C8 A0A2M4ACP7 Q29F06 A0A1I8P6Q8 A0A0Q9XE21 A0A1J1HP19 B4KXR0 D3TQ23 A0A1A9ZFZ1 T1H6F0 A0A1A9WTJ2 A0A1I8N3J9 A0A1A9V302 A0A1A9YL22 A0A1B0AR97 B4J1S3 E2BDI4 A0A0L0C6G6 E2A8F9 A0A195D4H0 A0A158NHF4 F4WD88 A0A195AZY6 A0A151XFD8 A0A067QZY2 E0VLJ1 A0A2J7RI51 A0A195FW16 A0A154PBT7 A0A195E231 K7IZ54 A0A088ARN8 A0A0L7RD18 B4LFP1 A0A310SI54 A0A2A3ELG3 A0A232EUD3 A0A0C9RQW4 A0A3L8DHD7 A0A1S3CU04 A0A0N0U4U7 A0A1W4WXL8 A0A2P8YLW5 A0A224XQJ9 R4G308 D2A5X1 A0A0A9ZGB8 A0A146KUB5 A0A0P6H904 A0A0P5ZSJ2
PDB
2F9F
E-value=0.0055571,
Score=94
Ontologies
PATHWAY
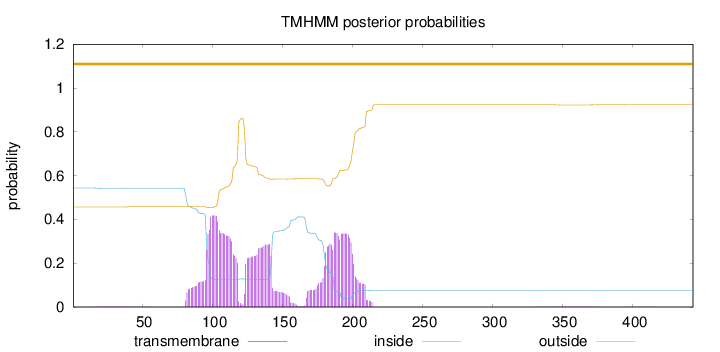
Topology
Length:
443
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
23.69194
Exp number, first 60 AAs:
0.0354
Total prob of N-in:
0.54304
outside
1 - 443
Population Genetic Test Statistics
Pi
177.837303
Theta
137.103215
Tajima's D
0.920226
CLR
0.271684
CSRT
0.642417879106045
Interpretation
Uncertain
