Gene
KWMTBOMO08225
Pre Gene Modal
BGIBMGA009503
Annotation
PREDICTED:_uncharacterized_protein_LOC101740576_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.819
Sequence
CDS
ATGATAGTGAAACTGAAAGAGGACCGGACGATCCTCCTCACGACGCATCATCTCGACGAAGCGGAGTTGCTAAGCGACCAAATCGTCATCATGCATAAGGGCCAAGTCCACACAACAGGCTCTCCGGTGGAGATAAAACGCACCCTGGGCAACGGGTACAAGCTAACCGTGGTCTTCCCCGAGACGACCTGCGAAGATCGCCCTGAAGACCTGGATGAGAAGACCAAACAACTGCTCTCTACCGTCAGGGACGACGTCAAGAACGCCAACATCGTCGATGTTAACGACACTGATGTGGAGATTCTACTGCCTTTCTATGACGCTGACGGACTTAGCAATGACCTACATCGCCTATGCTTGAACTTGGAGTCCCAGCAGCCAGTCCTCGGCTATAAGACCTACGCCATGGACTGCAGCAGTCTCGAGCAGGTGTTCTGTGACATCTGCACTCAAGCCGACCTAGGAGCATCTCAGACTGACGAGTCGAAATACGTATCCATAGAGTCGCCGTCTGAAGCGTCCGCCAGCACGAGGTCCCGTGGGGGGCGGGCGGGGGGCGCGGTGCGGGGGGCGGTGCGCGGGGAGGGCGCCCTGCGAGCCTCGTGCCGGGAGATGTTCGCGGCCCTCATGTACGCGCGGTACTTGCATCACGTCCGAAATAGATGGTTAATGTTCATGTTACTACTCCTGCCGTCGGTTTTCGTAACAGTAGCTATGGGATTCTCGTCTTTAAGACCGCCGGCTGTCAAAGAAATAGCTCTAAAATTAAATGCAGACCTGTACGAAGGATCTACGCAGTATATTGTGCTGCAGCCGTCTACGTATGAGAACAACGACGAAAAGTACGTAGGGTTCGGCGTTGACGTCCTCCAGAACATAGCTGGCGGTAACAACGTGACCACGTGGACTAGTAAGGACAATCCTTCGTGCACGTGCAACATAACGCGGCAGATCTGCGACGGAGACCCGGCTGCGGTCAGCACGCTGGCCATGATGGCGCTACCTGATGTGGATACACTCAACGAATGGCTGCTGACTTCACACGAAATATACATTGAGAAGAGATACGGCGGCTTCTCGTCGACAGTCAGAGACAACAAGACAAACTTCATAACGTGGTACAACAACAAGGGTCATCATGCGCTACCGTCATATCTCAACGTCATGAACAACGCCATCTATCGGACGGTGAGCGGTAACCGTCGTGCAAACATCACAGTATACACACATCCGTTGAAGATATCCAGTGAAAAGATCAACAAAGCCACTTTGTATCAGCACATAGCAGACGCTGGCATATCGAGCCTGGTGCTGGTGGCGTACGGGCTGGTGAGCGCGGGCGCGGGCGTGTGGCTGGTGGCGGCGCGCACCTCGCAGCAGAAGCGGCTCCAGCTACTCTGCGGAGTCAGCCCACTAATGTACTGGACCGCTGCACTCACTTGGGATGTCATTATCATCTTCATAAATATGATAATAACGGCGATGGTAATGTTGGCGTTCGGCTTCCCCGTGTTCGTGGCGAAAAACAACTTGCCGGCGATATGCGTACTCATTATGTTATATGGATTTGCGTGCGGTAACCTCGTCCATCTGTTGGAGAAGCTGTTTACGGAAGCTAGCCTAGCCAACATGGTGTTGTTCTGCGCGAATTCCTTGCTGGGGCTCGCCGGGATGACAATACTTCTCATACTCGACATCATATCGGATTCCGACGCAACAGACAACGCTCGGTGGATCTTACACAAGATATTCTTGATAGCTCCACAGTTCGCGCTGGGTGACGGATTACTGGAAATAGCCAAGAACACTATTCAGTCGGAGGTGTTAAGCCGTTTCGGTATGGACACGTATCAAGATCCGTTCAGGTCCAGTTTGCTAAGACTCCACGTGTTGGCGCTGGTAGTGGTAGGCGGAGCCCTCTTCCTACTCAACCTGGCCATCGAGTACGATTGCTTCGAAATATTTCTTGGCAGGTTGAGACGCGAGAAGATGCCGCAGACGAACTACGAAGAATTGGAGCCCGTGGAAGTCGCTGAGGAGAGGAAGAAGGTGCGAGCGGCACTGTTGCACTCCGCTCAGGGACACATCCGGGTCAACACCATCGGCAATATTAATAGGGGCTACGTACATACGGAGTGCGACGGCAAGCGAGTGCGGCGGGCGGCGCGTGGCGGCGGCGCCGGCGTGGCGCAATGTGTGCGGCTCGGCAAGATGTACCGCCGCCCCGCGCTCGTGGACCTCACGCTCTCGCTGCCTGAGGGACAGTGCACAGCGTTACTCGGAGAGAACGGCGCTGGTAAATCCACGACCTTCTCGATACTGACCGGGGAGATCCGACCTACTGCCGGTGAAGTGTACCTCAATGAACAGAAGATGACCCCTAGGAAACTCTGTCAAGGACTTATAAGTTACTGTCCACAAAGCGACGCTATAGACCCCCTGCTGACGGTCAGAGAGATCCTGTCCTTCTACTGCCAGTTAAGAGGAATAACTGACCAGGACGATGTGATAAAAAGAACATTAGCGATGTTTGAATTGAATTCATACAGTGACGTGCTGAGTGGAAATCTTAGTGGCGGAAACAAAAGGAAGTTGTGCACCGCTATTGCGTTCATGGGAAGAAATCCGCTTGTCCTACTCGATGAGCCGACCAGCGGTATGGACGCGTCGTCGGGCGCGGCGGTGTGCGAGCGGGCGCGCGAGTGCGCGGGCGAGGGCCGCGCCGTGCTCGTGGCCACACACGCGCTGCGGGACGCGCGACGTCTGGCCGCGCGCATCGCACTCCTGCGGAAGGGACGCCTCGTGGCTCTAGCGCCGCTACACGACTGCCTACAGAGGTTCGGCGGCGGGCACGTGGTGGAGTGCCGCGCGCCGGGAGCGGCGGGGGTCGCGTGGGGGACCGTGCGTGCTCGCGCCCCCTACGCCGTGCTGCGCGTGCTGCACGCACACACGCTGCAGTTCCTGCTACCCAACTACGTTACAGTTAACGAGAAGGAGACACCGATACGACTCAGCGATATATTCCGTTTGATGAACGAACTGCAGACTGCCGGTGAAATTGAGGACTACACAGTGAACCAGAGCTCGCTGGAACAGATGTTCCTAAGCTTCACGGACCAGAGGGATGTAGAAACGGGTCCCATCGAGATGGAACCTTTGCCAACGCCCGAATTTGTCAAGAGTGAAGAAGAATATGACAGCATCACCTGTCTGTAA
Protein
MIVKLKEDRTILLTTHHLDEAELLSDQIVIMHKGQVHTTGSPVEIKRTLGNGYKLTVVFPETTCEDRPEDLDEKTKQLLSTVRDDVKNANIVDVNDTDVEILLPFYDADGLSNDLHRLCLNLESQQPVLGYKTYAMDCSSLEQVFCDICTQADLGASQTDESKYVSIESPSEASASTRSRGGRAGGAVRGAVRGEGALRASCREMFAALMYARYLHHVRNRWLMFMLLLLPSVFVTVAMGFSSLRPPAVKEIALKLNADLYEGSTQYIVLQPSTYENNDEKYVGFGVDVLQNIAGGNNVTTWTSKDNPSCTCNITRQICDGDPAAVSTLAMMALPDVDTLNEWLLTSHEIYIEKRYGGFSSTVRDNKTNFITWYNNKGHHALPSYLNVMNNAIYRTVSGNRRANITVYTHPLKISSEKINKATLYQHIADAGISSLVLVAYGLVSAGAGVWLVAARTSQQKRLQLLCGVSPLMYWTAALTWDVIIIFINMIITAMVMLAFGFPVFVAKNNLPAICVLIMLYGFACGNLVHLLEKLFTEASLANMVLFCANSLLGLAGMTILLILDIISDSDATDNARWILHKIFLIAPQFALGDGLLEIAKNTIQSEVLSRFGMDTYQDPFRSSLLRLHVLALVVVGGALFLLNLAIEYDCFEIFLGRLRREKMPQTNYEELEPVEVAEERKKVRAALLHSAQGHIRVNTIGNINRGYVHTECDGKRVRRAARGGGAGVAQCVRLGKMYRRPALVDLTLSLPEGQCTALLGENGAGKSTTFSILTGEIRPTAGEVYLNEQKMTPRKLCQGLISYCPQSDAIDPLLTVREILSFYCQLRGITDQDDVIKRTLAMFELNSYSDVLSGNLSGGNKRKLCTAIAFMGRNPLVLLDEPTSGMDASSGAAVCERARECAGEGRAVLVATHALRDARRLAARIALLRKGRLVALAPLHDCLQRFGGGHVVECRAPGAAGVAWGTVRARAPYAVLRVLHAHTLQFLLPNYVTVNEKETPIRLSDIFRLMNELQTAGEIEDYTVNQSSLEQMFLSFTDQRDVETGPIEMEPLPTPEFVKSEEEYDSITCL
Summary
Uniprot
H9JJ02
A0A2H1VS40
A0A2W1BG17
A0A194PY81
A0A212EKX0
A0A151IL44
+ More
A0A1E1XFS1 A0A1S3KER8 A0A3S3Q091 A0A1V9X3M8 Q7PPR5 A0A453Z0G8 A0A182V7H8 A0A182X1U2 A0A182P8G6 A0A182HFQ7 A0A084WI53 B0WG45 A0A182MPU9 A0A182MYE2 A0A182JA80 A0A182K9A6 A0A182Q710 A0A182SLQ4 A0A182YA40 A0A182WCU7 A0A182UC32 A0A182LQ31 A0A369S9Q9 A0A3Q1ANV7 A0A3Q2PPB9 A0A3Q1CP84 A0A1W4VJ36 K1PG28 A0A3B4ZBY1 A0A3B0J768 A0A1B0CFQ3 B4R3P9 A0A1A8SFR3 B4PZ10 B3NYB4 A0A0Q5TLA7 B4I6E6 A0A1W5APQ7
A0A1E1XFS1 A0A1S3KER8 A0A3S3Q091 A0A1V9X3M8 Q7PPR5 A0A453Z0G8 A0A182V7H8 A0A182X1U2 A0A182P8G6 A0A182HFQ7 A0A084WI53 B0WG45 A0A182MPU9 A0A182MYE2 A0A182JA80 A0A182K9A6 A0A182Q710 A0A182SLQ4 A0A182YA40 A0A182WCU7 A0A182UC32 A0A182LQ31 A0A369S9Q9 A0A3Q1ANV7 A0A3Q2PPB9 A0A3Q1CP84 A0A1W4VJ36 K1PG28 A0A3B4ZBY1 A0A3B0J768 A0A1B0CFQ3 B4R3P9 A0A1A8SFR3 B4PZ10 B3NYB4 A0A0Q5TLA7 B4I6E6 A0A1W5APQ7
Pubmed
EMBL
BABH01023103
BABH01023104
ODYU01003815
SOQ43044.1
KZ150088
PZC73728.1
+ More
KQ459585 KPI98296.1 AGBW02014194 OWR42117.1 KQ977135 KYN05470.1 GFAC01001130 JAT98058.1 NCKU01001693 RWS11467.1 MNPL01026418 OQR67993.1 AAAB01008933 EAA09973.5 APCN01005565 ATLV01023913 KE525347 KFB49897.1 DS231923 EDS26741.1 AXCM01003055 AXCN02001048 NOWV01000035 RDD43655.1 JH816926 EKC20528.1 OUUW01000003 SPP78064.1 AJWK01010254 CM000366 EDX18517.1 HAEI01014598 SBS17067.1 CM000162 EDX03071.2 CH954180 EDV47593.2 KQS30565.1 KQS30566.1 CH480823 EDW56352.1
KQ459585 KPI98296.1 AGBW02014194 OWR42117.1 KQ977135 KYN05470.1 GFAC01001130 JAT98058.1 NCKU01001693 RWS11467.1 MNPL01026418 OQR67993.1 AAAB01008933 EAA09973.5 APCN01005565 ATLV01023913 KE525347 KFB49897.1 DS231923 EDS26741.1 AXCM01003055 AXCN02001048 NOWV01000035 RDD43655.1 JH816926 EKC20528.1 OUUW01000003 SPP78064.1 AJWK01010254 CM000366 EDX18517.1 HAEI01014598 SBS17067.1 CM000162 EDX03071.2 CH954180 EDV47593.2 KQS30565.1 KQS30566.1 CH480823 EDW56352.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000078542
UP000085678
UP000285301
+ More
UP000192247 UP000007062 UP000075903 UP000076407 UP000075885 UP000075840 UP000030765 UP000002320 UP000075883 UP000075884 UP000075880 UP000075881 UP000075886 UP000075901 UP000076408 UP000075920 UP000075902 UP000075882 UP000253843 UP000257160 UP000265000 UP000192221 UP000005408 UP000261400 UP000268350 UP000092461 UP000000304 UP000002282 UP000008711 UP000001292 UP000192224
UP000192247 UP000007062 UP000075903 UP000076407 UP000075885 UP000075840 UP000030765 UP000002320 UP000075883 UP000075884 UP000075880 UP000075881 UP000075886 UP000075901 UP000076408 UP000075920 UP000075902 UP000075882 UP000253843 UP000257160 UP000265000 UP000192221 UP000005408 UP000261400 UP000268350 UP000092461 UP000000304 UP000002282 UP000008711 UP000001292 UP000192224
PRIDE
Interpro
IPR003593
AAA+_ATPase
+ More
IPR026082 ABCA
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR017871 ABC_transporter_CS
IPR020846 MFS_dom
IPR003508 CIDE-N_dom
IPR018056 Kringle_CS
IPR013806 Kringle-like
IPR000001 Kringle
IPR038178 Kringle_sf
IPR030369 ABCA7
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR000515 MetI-like
IPR026082 ABCA
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR017871 ABC_transporter_CS
IPR020846 MFS_dom
IPR003508 CIDE-N_dom
IPR018056 Kringle_CS
IPR013806 Kringle-like
IPR000001 Kringle
IPR038178 Kringle_sf
IPR030369 ABCA7
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR000515 MetI-like
Gene 3D
ProteinModelPortal
H9JJ02
A0A2H1VS40
A0A2W1BG17
A0A194PY81
A0A212EKX0
A0A151IL44
+ More
A0A1E1XFS1 A0A1S3KER8 A0A3S3Q091 A0A1V9X3M8 Q7PPR5 A0A453Z0G8 A0A182V7H8 A0A182X1U2 A0A182P8G6 A0A182HFQ7 A0A084WI53 B0WG45 A0A182MPU9 A0A182MYE2 A0A182JA80 A0A182K9A6 A0A182Q710 A0A182SLQ4 A0A182YA40 A0A182WCU7 A0A182UC32 A0A182LQ31 A0A369S9Q9 A0A3Q1ANV7 A0A3Q2PPB9 A0A3Q1CP84 A0A1W4VJ36 K1PG28 A0A3B4ZBY1 A0A3B0J768 A0A1B0CFQ3 B4R3P9 A0A1A8SFR3 B4PZ10 B3NYB4 A0A0Q5TLA7 B4I6E6 A0A1W5APQ7
A0A1E1XFS1 A0A1S3KER8 A0A3S3Q091 A0A1V9X3M8 Q7PPR5 A0A453Z0G8 A0A182V7H8 A0A182X1U2 A0A182P8G6 A0A182HFQ7 A0A084WI53 B0WG45 A0A182MPU9 A0A182MYE2 A0A182JA80 A0A182K9A6 A0A182Q710 A0A182SLQ4 A0A182YA40 A0A182WCU7 A0A182UC32 A0A182LQ31 A0A369S9Q9 A0A3Q1ANV7 A0A3Q2PPB9 A0A3Q1CP84 A0A1W4VJ36 K1PG28 A0A3B4ZBY1 A0A3B0J768 A0A1B0CFQ3 B4R3P9 A0A1A8SFR3 B4PZ10 B3NYB4 A0A0Q5TLA7 B4I6E6 A0A1W5APQ7
PDB
5XJY
E-value=1.90768e-42,
Score=438
Ontologies
GO
PANTHER
Topology
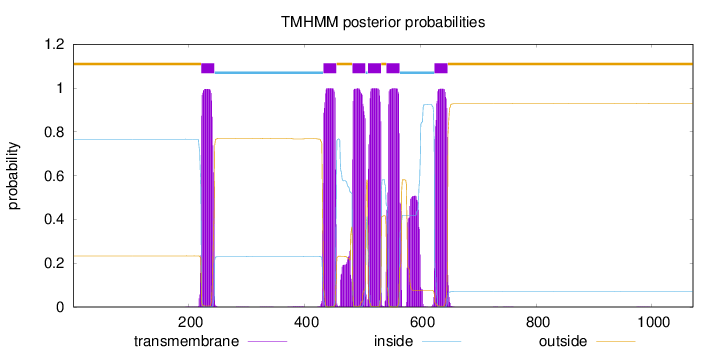
Length:
1071
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
149.141250000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.76659
outside
1 - 221
TMhelix
222 - 244
inside
245 - 432
TMhelix
433 - 455
outside
456 - 482
TMhelix
483 - 505
inside
506 - 509
TMhelix
510 - 532
outside
533 - 541
TMhelix
542 - 564
inside
565 - 624
TMhelix
625 - 647
outside
648 - 1071
Population Genetic Test Statistics
Pi
258.042721
Theta
188.577114
Tajima's D
1.552666
CLR
0.190741
CSRT
0.793060346982651
Interpretation
Uncertain
