Gene
KWMTBOMO08216
Annotation
neuropeptide_receptor_A27_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.767
Sequence
CDS
ATGCACCGGACGTCTTATCCTTTGGGCCACGACGACTACTACTGGATTATTATGCGCCACTCCTGCCTATCCTCGGCTGTGTGGATATCAACGTGGACTGAAGTAGCCTCGTACGGTTCGGTGCTGACCATCGTCGCGTTCTCCCTGGAGCGCTACCTAGCTATCTGCCACCCGCTCCACTTGTACGCGATGGCTGGACTGCGCCGCGCCCTCAGGGTGGTGGCCGCCCTGTGGCTGCTGTCCTTTGTCGCGGCAGCTCCGTTCGCGTCGTATACCACGGTCAGCTACCACGATTACCCTCCAGGTTCCGGAAACGCTTCTCTAGAGTCAGCATTCTGCGCGATGCTGGAAGTACCCTCGTGGTACCTCTACGAGCTGAGCAGCCTCCTGTTCTTCATCCTCCCTGGGCTGATAATACTCTGTCTTTACGTCAGGATGGGACTCCGGATCAGGTATGTTATGGGGTACGTATCTATCTTCTACCTACCTCTACTTCTCTACCTATTTCTACATTAA
Protein
MHRTSYPLGHDDYYWIIMRHSCLSSAVWISTWTEVASYGSVLTIVAFSLERYLAICHPLHLYAMAGLRRALRVVAALWLLSFVAAAPFASYTTVSYHDYPPGSGNASLESAFCAMLEVPSWYLYELSSLLFFILPGLIILCLYVRMGLRIRYVMGYVSIFYLPLLLYLFLH
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
A0A2H1VQB5
A0A0S1YDP4
A0A0N1PIL2
A0A0C9RF21
E0VKF7
U3U7N4
+ More
A0A158P1A1 A0A195BSD8 A0A195F3A2 A0A151J3I9 A0A195CID3 A0A151X912 F4W9B0 A0A195F324 A0A195CIE0 E9IKY0 A0A1S4EN32 A0A026W1H8 A0A3L8D7M4 A0A151J3W6 A0A2S2PNY7 A0A195BT10 A0A158P193 A0A3L8D852 E2BGR8 A0A026W2V1 J9K383 E2AQX8 A0A2A3ERA9 A0A1B6C172 D6P3E6 A0A067RG65 E2BGR7 T1H917 D6P3E5 A1X4Q3 A0A2J7QR84 A0A154NZ86 A0A1L8DJI0 A0A1L8DJT5 A0A067QYB7 A0A336L8B3 A0A336MKH8 A0A154NXN6 A0A146M155 A0A182TGB0 A0A182PE69 A0A182JY61 A0A182SU37 A0A0C9RTW4 A0A182N583 A0A182M1V0 A0A182VQQ1 A0A2C9H888 A0A182YKD8 A0A1J1IM35 A7XII0 A0A182JGG2 A0A3F2Z0D9 A0A1S4GHC2 A0A182LCK4 A0A2Y9D0Z8 A0A2C9GWQ3 A0A1B6LAI1 J9HXV9 A0A1B6EVK3 A0A182FSI7 A0A0P5LFX8 A0A2C9GW61 A0A0P4XFK5 A0A0P5TEF7 E9H1R2 B4MXQ7 A0A1Y1NI26 A0A411JKC9 A0A0P4XR87 A0A0N8E0K3 A0A0N7ZQR3 A0A0P5HFY0 A0A0N8E807 A0A0P5YF00
A0A158P1A1 A0A195BSD8 A0A195F3A2 A0A151J3I9 A0A195CID3 A0A151X912 F4W9B0 A0A195F324 A0A195CIE0 E9IKY0 A0A1S4EN32 A0A026W1H8 A0A3L8D7M4 A0A151J3W6 A0A2S2PNY7 A0A195BT10 A0A158P193 A0A3L8D852 E2BGR8 A0A026W2V1 J9K383 E2AQX8 A0A2A3ERA9 A0A1B6C172 D6P3E6 A0A067RG65 E2BGR7 T1H917 D6P3E5 A1X4Q3 A0A2J7QR84 A0A154NZ86 A0A1L8DJI0 A0A1L8DJT5 A0A067QYB7 A0A336L8B3 A0A336MKH8 A0A154NXN6 A0A146M155 A0A182TGB0 A0A182PE69 A0A182JY61 A0A182SU37 A0A0C9RTW4 A0A182N583 A0A182M1V0 A0A182VQQ1 A0A2C9H888 A0A182YKD8 A0A1J1IM35 A7XII0 A0A182JGG2 A0A3F2Z0D9 A0A1S4GHC2 A0A182LCK4 A0A2Y9D0Z8 A0A2C9GWQ3 A0A1B6LAI1 J9HXV9 A0A1B6EVK3 A0A182FSI7 A0A0P5LFX8 A0A2C9GW61 A0A0P4XFK5 A0A0P5TEF7 E9H1R2 B4MXQ7 A0A1Y1NI26 A0A411JKC9 A0A0P4XR87 A0A0N8E0K3 A0A0N7ZQR3 A0A0P5HFY0 A0A0N8E807 A0A0P5YF00
Pubmed
EMBL
ODYU01003784
SOQ42976.1
KT031025
ALM88323.1
KQ460889
KPJ11065.1
+ More
GBYB01005586 JAG75353.1 DS235243 EEB13873.1 AB817308 BAO01075.1 ADTU01001054 ADTU01001055 KQ976418 KYM89502.1 KQ981855 KYN34861.1 KQ980275 KYN17015.1 KQ977720 KYN00490.1 KQ982402 KYQ56852.1 GL888002 EGI69292.1 KYN34863.1 KYN00488.1 GL764026 EFZ18684.1 MG550214 AWT50648.1 KK107488 EZA49917.1 QOIP01000012 RLU16264.1 KYN17017.1 GGMR01018514 MBY31133.1 KYM89503.1 ADTU01001049 ADTU01001050 ADTU01001051 RLU16263.1 GL448228 EFN85050.1 EZA49916.1 ABLF02039665 ABLF02039672 ABLF02039675 ABLF02039682 ABLF02039685 ABLF02045610 GL441846 EFN64124.1 KZ288192 PBC34305.1 GEDC01030042 JAS07256.1 GU734128 ADG27753.1 KK852492 KDR22757.1 EFN85049.1 ACPB03001653 GU734127 ADG27752.1 DQ201783 ABB05503.1 NEVH01011922 PNF31102.1 KQ434778 KZC04374.1 GFDF01007478 JAV06606.1 GFDF01007474 JAV06610.1 KK852830 KDR15327.1 UFQS01001286 UFQT01001286 SSX10125.1 SSX29847.1 UFQT01001318 SSX30011.1 KZC04373.1 GDHC01005085 JAQ13544.1 GBYB01011021 JAG80788.1 AXCM01004259 AXCM01004260 CVRI01000051 CRK99529.1 AY900217 AAX84796.1 AAAB01008879 APCN01002146 APCN01002147 GEBQ01032227 GEBQ01019322 GEBQ01014855 GEBQ01008807 GEBQ01004738 JAT07750.1 JAT20655.1 JAT25122.1 JAT31170.1 JAT35239.1 CH477186 EJY57320.1 GECZ01027831 JAS41938.1 GDIQ01172055 JAK79670.1 AXCN02000164 GDIP01243398 JAI80003.1 GDIP01127636 JAL76078.1 GL732584 EFX74323.1 CH963876 EDW76826.2 GEZM01007119 JAV95367.1 MH810205 QBC65470.1 GDIP01238781 JAI84620.1 GDIQ01073644 JAN21093.1 GDIP01221833 JAJ01569.1 GDIQ01230240 JAK21485.1 GDIQ01052812 JAN41925.1 GDIP01058794 JAM44921.1
GBYB01005586 JAG75353.1 DS235243 EEB13873.1 AB817308 BAO01075.1 ADTU01001054 ADTU01001055 KQ976418 KYM89502.1 KQ981855 KYN34861.1 KQ980275 KYN17015.1 KQ977720 KYN00490.1 KQ982402 KYQ56852.1 GL888002 EGI69292.1 KYN34863.1 KYN00488.1 GL764026 EFZ18684.1 MG550214 AWT50648.1 KK107488 EZA49917.1 QOIP01000012 RLU16264.1 KYN17017.1 GGMR01018514 MBY31133.1 KYM89503.1 ADTU01001049 ADTU01001050 ADTU01001051 RLU16263.1 GL448228 EFN85050.1 EZA49916.1 ABLF02039665 ABLF02039672 ABLF02039675 ABLF02039682 ABLF02039685 ABLF02045610 GL441846 EFN64124.1 KZ288192 PBC34305.1 GEDC01030042 JAS07256.1 GU734128 ADG27753.1 KK852492 KDR22757.1 EFN85049.1 ACPB03001653 GU734127 ADG27752.1 DQ201783 ABB05503.1 NEVH01011922 PNF31102.1 KQ434778 KZC04374.1 GFDF01007478 JAV06606.1 GFDF01007474 JAV06610.1 KK852830 KDR15327.1 UFQS01001286 UFQT01001286 SSX10125.1 SSX29847.1 UFQT01001318 SSX30011.1 KZC04373.1 GDHC01005085 JAQ13544.1 GBYB01011021 JAG80788.1 AXCM01004259 AXCM01004260 CVRI01000051 CRK99529.1 AY900217 AAX84796.1 AAAB01008879 APCN01002146 APCN01002147 GEBQ01032227 GEBQ01019322 GEBQ01014855 GEBQ01008807 GEBQ01004738 JAT07750.1 JAT20655.1 JAT25122.1 JAT31170.1 JAT35239.1 CH477186 EJY57320.1 GECZ01027831 JAS41938.1 GDIQ01172055 JAK79670.1 AXCN02000164 GDIP01243398 JAI80003.1 GDIP01127636 JAL76078.1 GL732584 EFX74323.1 CH963876 EDW76826.2 GEZM01007119 JAV95367.1 MH810205 QBC65470.1 GDIP01238781 JAI84620.1 GDIQ01073644 JAN21093.1 GDIP01221833 JAJ01569.1 GDIQ01230240 JAK21485.1 GDIQ01052812 JAN41925.1 GDIP01058794 JAM44921.1
Proteomes
UP000053240
UP000009046
UP000005205
UP000078540
UP000078541
UP000078492
+ More
UP000078542 UP000075809 UP000007755 UP000079169 UP000053097 UP000279307 UP000008237 UP000007819 UP000000311 UP000242457 UP000027135 UP000015103 UP000235965 UP000076502 UP000075902 UP000075885 UP000075881 UP000075901 UP000075884 UP000075883 UP000075920 UP000076407 UP000076408 UP000183832 UP000075880 UP000075903 UP000075882 UP000075840 UP000075900 UP000008820 UP000069272 UP000075886 UP000000305 UP000007798
UP000078542 UP000075809 UP000007755 UP000079169 UP000053097 UP000279307 UP000008237 UP000007819 UP000000311 UP000242457 UP000027135 UP000015103 UP000235965 UP000076502 UP000075902 UP000075885 UP000075881 UP000075901 UP000075884 UP000075883 UP000075920 UP000076407 UP000076408 UP000183832 UP000075880 UP000075903 UP000075882 UP000075840 UP000075900 UP000008820 UP000069272 UP000075886 UP000000305 UP000007798
Interpro
SUPFAM
SSF47095
SSF47095
ProteinModelPortal
A0A2H1VQB5
A0A0S1YDP4
A0A0N1PIL2
A0A0C9RF21
E0VKF7
U3U7N4
+ More
A0A158P1A1 A0A195BSD8 A0A195F3A2 A0A151J3I9 A0A195CID3 A0A151X912 F4W9B0 A0A195F324 A0A195CIE0 E9IKY0 A0A1S4EN32 A0A026W1H8 A0A3L8D7M4 A0A151J3W6 A0A2S2PNY7 A0A195BT10 A0A158P193 A0A3L8D852 E2BGR8 A0A026W2V1 J9K383 E2AQX8 A0A2A3ERA9 A0A1B6C172 D6P3E6 A0A067RG65 E2BGR7 T1H917 D6P3E5 A1X4Q3 A0A2J7QR84 A0A154NZ86 A0A1L8DJI0 A0A1L8DJT5 A0A067QYB7 A0A336L8B3 A0A336MKH8 A0A154NXN6 A0A146M155 A0A182TGB0 A0A182PE69 A0A182JY61 A0A182SU37 A0A0C9RTW4 A0A182N583 A0A182M1V0 A0A182VQQ1 A0A2C9H888 A0A182YKD8 A0A1J1IM35 A7XII0 A0A182JGG2 A0A3F2Z0D9 A0A1S4GHC2 A0A182LCK4 A0A2Y9D0Z8 A0A2C9GWQ3 A0A1B6LAI1 J9HXV9 A0A1B6EVK3 A0A182FSI7 A0A0P5LFX8 A0A2C9GW61 A0A0P4XFK5 A0A0P5TEF7 E9H1R2 B4MXQ7 A0A1Y1NI26 A0A411JKC9 A0A0P4XR87 A0A0N8E0K3 A0A0N7ZQR3 A0A0P5HFY0 A0A0N8E807 A0A0P5YF00
A0A158P1A1 A0A195BSD8 A0A195F3A2 A0A151J3I9 A0A195CID3 A0A151X912 F4W9B0 A0A195F324 A0A195CIE0 E9IKY0 A0A1S4EN32 A0A026W1H8 A0A3L8D7M4 A0A151J3W6 A0A2S2PNY7 A0A195BT10 A0A158P193 A0A3L8D852 E2BGR8 A0A026W2V1 J9K383 E2AQX8 A0A2A3ERA9 A0A1B6C172 D6P3E6 A0A067RG65 E2BGR7 T1H917 D6P3E5 A1X4Q3 A0A2J7QR84 A0A154NZ86 A0A1L8DJI0 A0A1L8DJT5 A0A067QYB7 A0A336L8B3 A0A336MKH8 A0A154NXN6 A0A146M155 A0A182TGB0 A0A182PE69 A0A182JY61 A0A182SU37 A0A0C9RTW4 A0A182N583 A0A182M1V0 A0A182VQQ1 A0A2C9H888 A0A182YKD8 A0A1J1IM35 A7XII0 A0A182JGG2 A0A3F2Z0D9 A0A1S4GHC2 A0A182LCK4 A0A2Y9D0Z8 A0A2C9GWQ3 A0A1B6LAI1 J9HXV9 A0A1B6EVK3 A0A182FSI7 A0A0P5LFX8 A0A2C9GW61 A0A0P4XFK5 A0A0P5TEF7 E9H1R2 B4MXQ7 A0A1Y1NI26 A0A411JKC9 A0A0P4XR87 A0A0N8E0K3 A0A0N7ZQR3 A0A0P5HFY0 A0A0N8E807 A0A0P5YF00
Ontologies
KEGG
GO
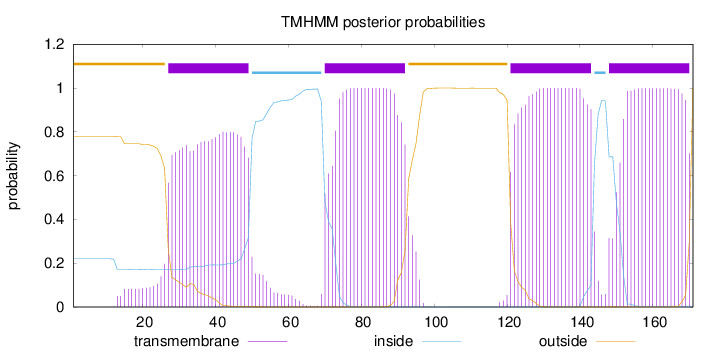
Topology
Length:
171
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
84.63163
Exp number, first 60 AAs:
19.5812
Total prob of N-in:
0.22029
POSSIBLE N-term signal
sequence
outside
1 - 26
TMhelix
27 - 49
inside
50 - 69
TMhelix
70 - 92
outside
93 - 120
TMhelix
121 - 143
inside
144 - 147
TMhelix
148 - 170
outside
171 - 171
Population Genetic Test Statistics
Pi
354.287564
Theta
204.405868
Tajima's D
2.157264
CLR
0.18498
CSRT
0.906954652267387
Interpretation
Uncertain
