Gene
KWMTBOMO08213 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009463
Annotation
Host_cell_factor_1_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.078
Sequence
CDS
ATGAAGGAAAACGCGGTACTTAAGTGGCAAAAAGTGTACAATCCTACCGGCCCTCAACCGCGACCCCGCCATGGCCATCGCGCGGTCGCCATAAAAGATTTGATGATAGTTTTCGGCGGCGGAAACGAAGGAATCGTTCACGAACTGCATGTCTTCAATACCACCACTAATCAGTGGTTCGTACCAGTCACAAAAGGAGAAGTCCCACCAGGATGCGCTGCCTACGGCTTCGTGGTAGACGGTACGCGCCTGTTAGTTTTTGGCGGCATGGTAGAGTACGGCAAGTATTCAAATGATTTGTACGAATTACAAGCCTCTAGGTGGGAATGGAAACGTTTAAAACCACTGCCACCAAAGCAGGGTCTTCCACCTTGTCCACGACTCGGTCACAGCTTTACGCTTCTCAATGGGAAAGTATACCTGTTTGGCGGACTGGCCAATGAAAGCGACGATCCAAAAAATAACATACCGAGATATCTTAATGATCTATACATTTTAGGACTTTATCCTAATTCATCTATGACTGTTTGGGATATACCTTTGACCTACGGTCAGTCTCCACCTCCCCGAGAGTCTCACAGTGGGGTAGCATATACTGACAAAAACACAGGCAAATCATCTTTAATTATCTATGGAGGCATGAGTGGATCTCGCCTTGGTGATTTGTGGATACTTGATGTTGACACAATGTCATGGTCTCGACCGGAAGTTGGAGGACCTCCACCTCTTCCTAGATCACTTCATACAGCTACAGTTATCGGCCACCACATGTATGTCTATGGTGGCTGGGTACCGCTGGTCCCTGATGAATCTAAACTTGCTACACATGAGAAAGAATGGAAGTGCACAAACACCCTTGCCTCATTGAACCTAGAAACAATGACTTGGGACAATATAGCACTGGACAAATTTGAGGAATGCGTTCCTCGGGCTCGAGCTGGACACAGTGCTGTTGCTATTCAAACTAGACTGTACATATGGTCTGGAAGGGATGGATATAGGAAAACTTGGAACAACCAAATCTGTTGCAAAGACCTTTGGTATCTTGAAGTAGGAGTACCACCACAAGCGGGACGTGTAGCCCTTGTACGTGCTGGCACCACATCATTGGAACTATGCTGGCCTGCTCTCAACACAGTTACCACTTATTTATTGCAAGTACAGAAATGTGGCAAAGTGACACCTACACGTTTCCCGGCTGCCTCTGAACCTGTGGTTGCACCTCCTCCTGCATCTCCTGCTGCCACACAGACTGATGCTGCTAAAGCTTTTGGTCTATCTTCTCCAGTTTCTTCATCGATCCCCATCACACCATCTGATACAAATGCTCGTGTAGTAGTTACAGTAGCTTCACCTATAGTGACACCAATCATCACGACTCCTCAAAAAGTAACACCTGGAACACTCAAACTGTCTGCTCAAGCTGCCACCGTAAAAGTCACGCCGGGCACTCCAACTACGAAACAGTCATTCCAGGCGAAAGCAATAGTCAAATCTCCCGCTAATCCTGCGACGCCTAGCTCCTCTCAACAGATCAAAGTGGCCGCGGTGACGCCGCAAGGTGTGACAAGAATCGTAAGTGGCGTCTCGACACCAGGAACCGTACGTGTCTCTACAGCACAGTCTGGTGCACAGATTGTGTTAGGATCGGCCTCTGGAGGCGCCGGTACGCCGCGTTACGTACAAGTGAAAACAGGCGGGAGTGGTGCTGTTCCTGTGAAGTTGAGTGCTGGTAATATGCCTGTCAAACTTGCAGGTAATGCAGTCCCACTAAAGATTGGTACTACTAACATTCAAGGAAAATTGACTCCCAACAGTGTGCAGAAAGTAGGACAAGTCACTGCTGGAAATCTACCCTTGAAACTTGGTGCCGGTAATGTCAAGATTGGTACGAGTAATGTTCTTGTAAAAACAGCAGGCGGAATGCCCTTACAGGGTGGTGTTGCAAACCTACAAACAAAAGTCGCCACCGGGGTTCCTGTGAAACTAGGCGCCGGTAACATTCCTGTGATAGCCAGCTCAGGGGGCACGATTCAAATAGCAGGTGGAACGCTGGCTGGCCAACAAGTAAAAACACCTGTATATAAGATATTGACAGCAAAATCAGGTGATCAAACAGCAACAGTGGCTACATCAACAGCAACTGGTGCGACAGTACTGAGGCAAGCAGCTGGCAATGCTGGTATTCCCGTCATCATAAAGAAAACAGGTGCAGGTGCGCAAGCAAGTAATACACCGCAATTTGTTACCCTTGTGAAGACCTCAACGGGCATGACTGTAGCAACTATGCCAAAAGTAGCAATGATGAACAGGTCTGGAGCAGCCGTTTCTCAAGCACAAAACATCACTCCGGGGGCCACTATTGTAAAGTTAGTATCTAACACTGTTGGTGGTAACAAAATTATCACTTTACCATCTAATATGTTGCAACTAGGTAAGGCTGGTGTTGGTGGAAAACAGACCATAGTAATCACCAAATCAGCTAGCCAAGCAGCACAGTCAAATCAACAACAGTGA
Protein
MKENAVLKWQKVYNPTGPQPRPRHGHRAVAIKDLMIVFGGGNEGIVHELHVFNTTTNQWFVPVTKGEVPPGCAAYGFVVDGTRLLVFGGMVEYGKYSNDLYELQASRWEWKRLKPLPPKQGLPPCPRLGHSFTLLNGKVYLFGGLANESDDPKNNIPRYLNDLYILGLYPNSSMTVWDIPLTYGQSPPPRESHSGVAYTDKNTGKSSLIIYGGMSGSRLGDLWILDVDTMSWSRPEVGGPPPLPRSLHTATVIGHHMYVYGGWVPLVPDESKLATHEKEWKCTNTLASLNLETMTWDNIALDKFEECVPRARAGHSAVAIQTRLYIWSGRDGYRKTWNNQICCKDLWYLEVGVPPQAGRVALVRAGTTSLELCWPALNTVTTYLLQVQKCGKVTPTRFPAASEPVVAPPPASPAATQTDAAKAFGLSSPVSSSIPITPSDTNARVVVTVASPIVTPIITTPQKVTPGTLKLSAQAATVKVTPGTPTTKQSFQAKAIVKSPANPATPSSSQQIKVAAVTPQGVTRIVSGVSTPGTVRVSTAQSGAQIVLGSASGGAGTPRYVQVKTGGSGAVPVKLSAGNMPVKLAGNAVPLKIGTTNIQGKLTPNSVQKVGQVTAGNLPLKLGAGNVKIGTSNVLVKTAGGMPLQGGVANLQTKVATGVPVKLGAGNIPVIASSGGTIQIAGGTLAGQQVKTPVYKILTAKSGDQTATVATSTATGATVLRQAAGNAGIPVIIKKTGAGAQASNTPQFVTLVKTSTGMTVATMPKVAMMNRSGAAVSQAQNITPGATIVKLVSNTVGGNKIITLPSNMLQLGKAGVGGKQTIVITKSASQAAQSNQQQ
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF01344 Kelch_1
SUPFAM
SSF117281
SSF117281
Gene 3D
ProteinModelPortal
PDB
6DO5
E-value=3.82998e-12,
Score=176
Ontologies
GO
Topology
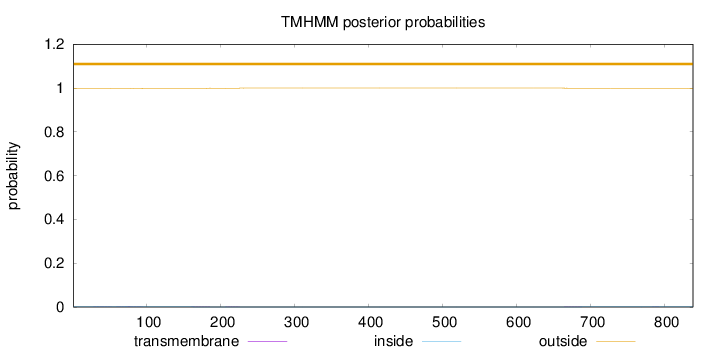
Length:
838
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0858100000000001
Exp number, first 60 AAs:
0.01309
Total prob of N-in:
0.00332
outside
1 - 838
Population Genetic Test Statistics
Pi
239.761558
Theta
185.666694
Tajima's D
0.987935
CLR
0
CSRT
0.655067246637668
Interpretation
Possibly Positive selection
